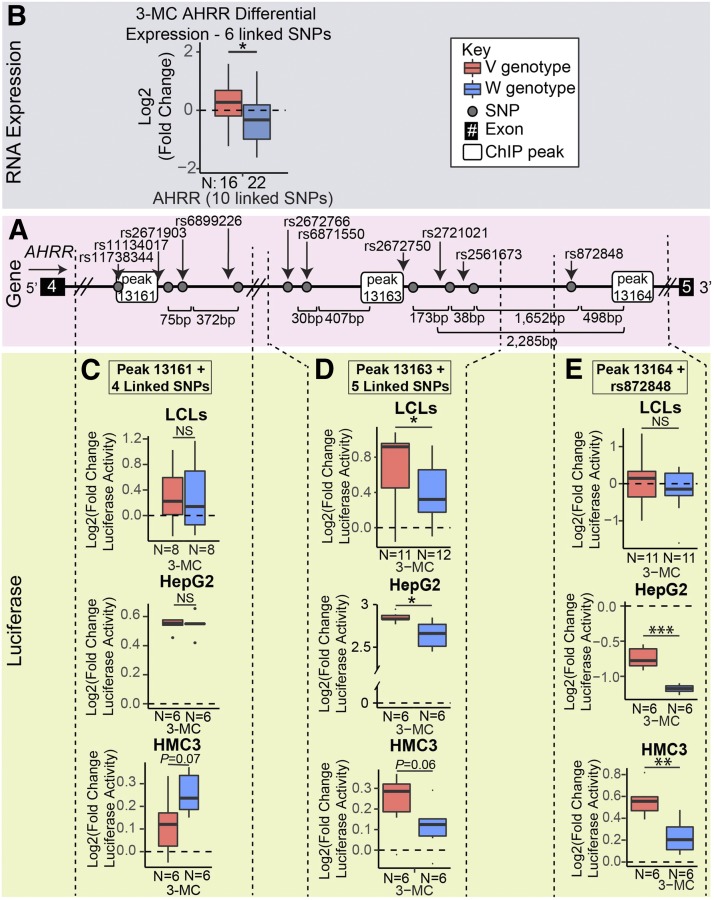
Fig. 6.
Functional investigation of the AHRR 3-MC–dependent PGx-eQTL. (A) The 3-MC–dependent PGx-eQTL for AHRR included 10 SNPs across peaks 13161, 13163, and 13164 in intron 4 of the AHRR gene. (B) The variant SNP genotypes for the 10 linked SNPs that were across three AHR-binding peaks in intron 4 of the AHRR gene were associated with an increase after 3-MC treatment, and the wild-type was associated with a decrease after 3-MC treatment. (C) The luciferase plasmid that contained the variant SNP genotypes for the SNPs surrounding peak 13161 (as indicated by the dashed lines) demonstrated no difference in LCLs or HepG2 cells and a smaller increase than the wild-type SNP genotype in HMC3 cells, which was suggestive. (D) The luciferase plasmid that contained the variant genotypes for the five SNPs near peak 13163 in intron 4 of the AHRR gene (as indicated by the dashed lines) demonstrated a larger increase in luciferase activity after 3-MC treatment than the wild-type in LCLs, HepG2, and HMC3—the same direction observed for differential expression in LCLs. (E) The luciferase plasmid that contained the variant genotype for rs872848 and peak 13164 (indicated by the dashed lines) demonstrated no difference in luciferase activity in LCLs, but showed a smaller decrease than did the plasmid that contained the wild-type genotype for rs872848 with peak 13164 in HepG2 and displayed a larger increase in luciferase activity in HMC3. The number of samples (N) for each group is at the bottom of each plot. All tests for significance used a two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.