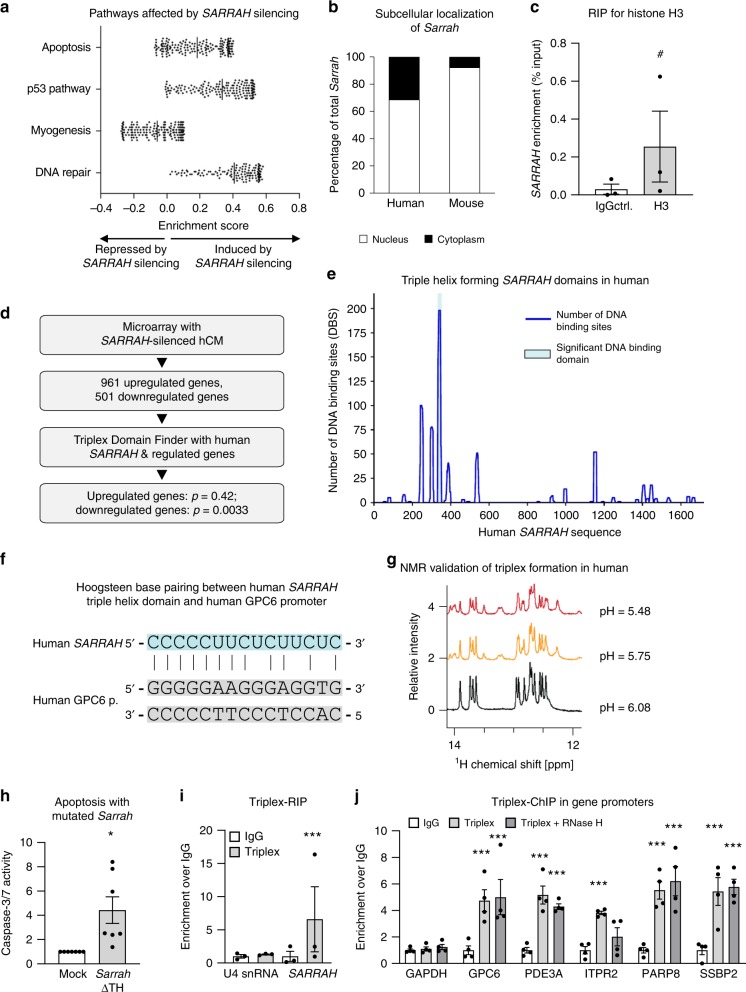
Fig. 3. SARRAHactivates gene expression via triple helix formation with gene promoters.
a Microarray data of primary human cardiomyocytes treated with GapmeR control or GapmeR SARRAH were analyzed for differentially regulated pathways using gene set enrichment analysis. b Human (primary) and mouse (HL-1 cell line) cardiomyocytes were fractionated into their cytoplasmic and nuclear portions and Sarrah levels were measured by qRT-PCR in both fractions (n = 3). c RNA-immunoprecipitation with an anti-total histone H3 antibody was performed in primary human cardiomyocytes. SARRAH levels were measured by qRT-PCR (n = 3; SEM; # t-test p = 0.0504; IgG immunoglobulin G). d Flow chart illustrating the procedure of microarray analysis of primary SARRAH-silenced human cardiomyocytes (hCM), identification of SARRAH DNA binding domains and DNA binding sites. e The human SARRAH sequence was assessed with regard to pyrimidine-rich regions capable of DNA binding via triple helix formation using the Triplex Domain Finder software. f Scheme indicating Hoogsteen base pairing between the human SARRAH triple helix domain and the human GPC6 promoter. g 1H spectra of the SARRAH binding site in the human GPC6 promoter as a DNA duplex 15mer in the presence of the human SARRAH triple helix domain as equal molar single-stranded RNA, as analyzed by nuclear magnetic resonance (NMR). h Using the CRISPR/Cas9-mediated approach outlined in Supplementary Fig. 7e the Sarrah triple helix domain was excised from the endogenous gene locus in mouse cardiomyocytes (HL-1 cell line). Apoptosis was quantified as caspase-3/7 activity (n = 7; SEM; *t-test p = 0.0156). i RNA-immunoprecipitation with the S9.6 anti-DNA-RNA-hybrid antibody was performed in crosslinked primary human cardiomyocytes. Levels of U4 snRNA as a negative control and SARRAH were measured by qRT-PCR (n = 3; SEM; ***t-test p = 0.0003; IgG immunoglobulin G). j Chromatin-immunoprecipitation with the S9.6 anti-DNA-RNA-hybrid antibody was performed in crosslinked primary human cardiomyocytes. Sonicated DNA fragments were used for qRT-PCR to quantify triplex formation in gene promoters (n = 4; SEM; ***two-way ANOVA IgG vs. RNA-DNA hybrid: p = 0.00017 for GPC6, p = 0.0044 for ITPR2 and p < 0.0001 for all other promoters; two-way ANOVA IgG vs. RNA-DNA hybrid + RNase H: p = 0.00089 for PDE3A and p < 0.0001 for all other promoters; F = 11.24 for variable “promoter” and F = 52.8 for variable “antibody”; IgG immunoglobulin G).