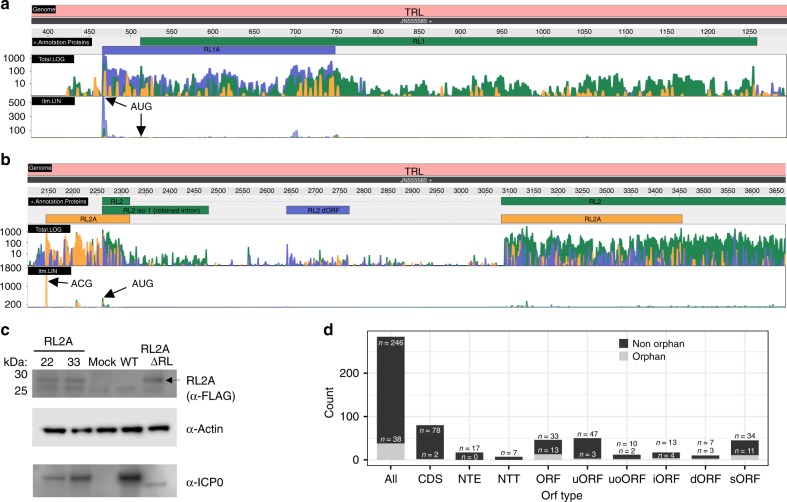
Fig. 6. Evidence of additional protein-coding sequences.
Ribosome profiling data visualizing expression of the two viral open reading frames (ORFs), RL1A and RL2A (the three colors depicting the read count for each of the three possible frames, yellow = 1, blue = 2, green = 3), expressed from the a RL1 and b RL2 locus of the terminal repeats (TRL). Both standard ribosome profiling data in log scale (Total.log) as well as translation start site profiling data obtained using Lactimidomycin (linear scale, ltm.LIN) are shown. The three possible reading frames are colored in yellow (frame 1), blue (frame 2) and green (frame 3). Both ORFs are well expressed and validated by the strong peak of their respective Translation start site (TaSS) in the ltm-track (black arrows). While RL2A initiates from a non-canonical ACG start codon, the 93 aa RL1A protein initiates from an AUG start codon and was previously missed due to its length of <100 aa. c Validation of RL2A expression by Western blot. Primary human fibroblasts were infected with two viral clones (22, 33) with one RL segment expressing a 3×-FLAG-tagged RL2A (RL2A), mock, wild-type HSV-1 (WT; for 24 h) or 3×-FLAG-RL2A-ΔRL (=RL2A-ΔRL) for 24 h. Interestingly, insertion of the 3×-FLAG-tag resulted in a loss of ICP0 expression presumably due to the introduction of three out-of-frame AUG start codons (within each FLAG-tag) upstream of the ICP0 TaSS. This was most pronounced when the second repeat was deleted (RL2A-ΔRL). Actin served as house-keeping control. A representative experiment of two independent experiments is shown. d Distributions of all identified types of ORFs (CDS coding sequence, NTE N-terminal extended ORFs, NTT N-terminal truncated ORFs, uORF upstream ORF, uoORF upstream overlapping ORF, iORF internal ORF, dORF downstream ORF, sORF short ORF) of HSV-1 classified by ORFs and orphan ORFs. Source data are provided within the Source Data file.