Figure 4. nonu-1-Dependent mRNA Cleavage Occurs in the Vicinity of Stalled Ribosomes.
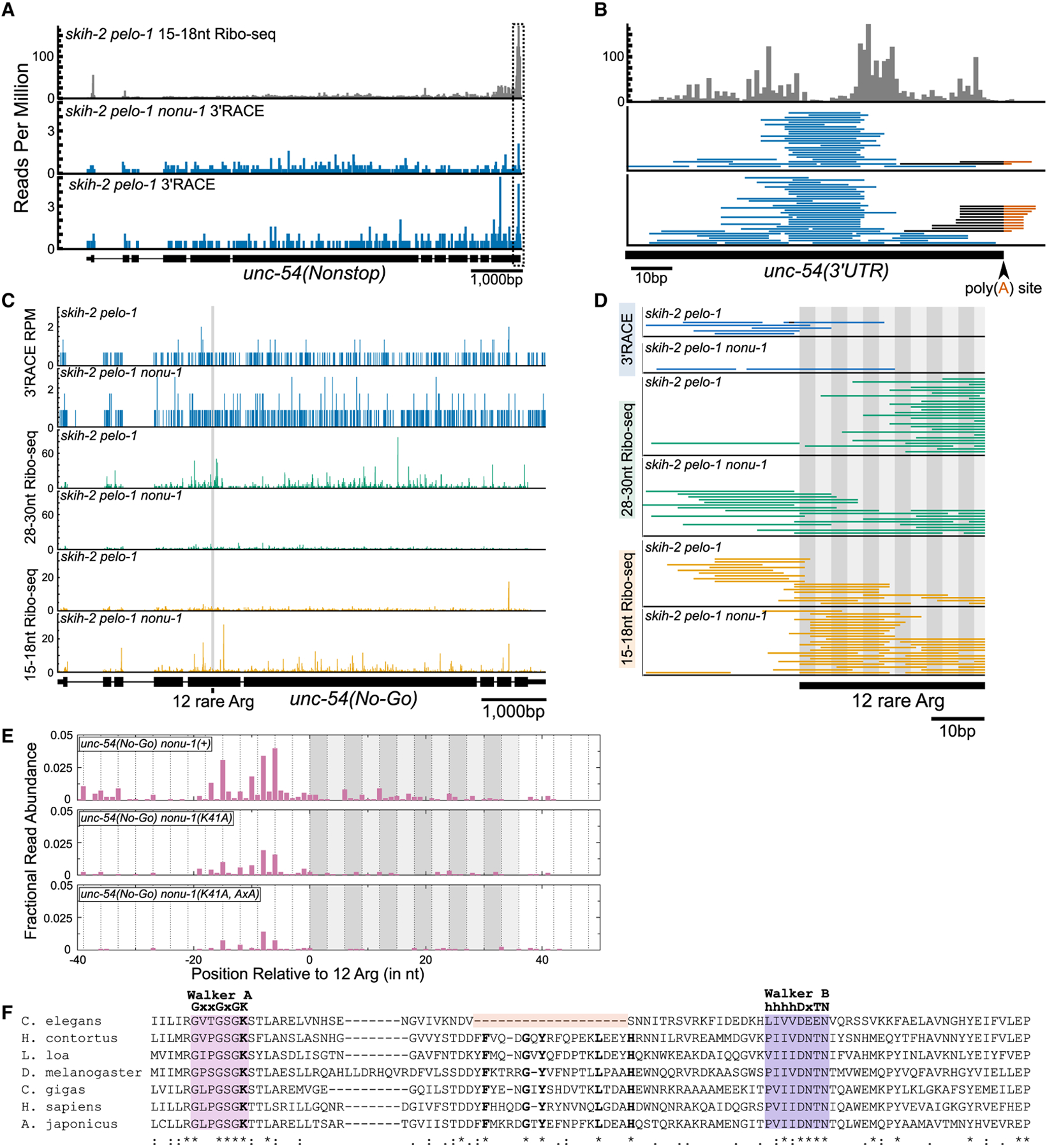
(A) 15–18 nt Ribo-seq reads from (Arribere and Fire, 2018) on top (gray) and 3′ RACE data from this study on bottom (blue). The 5′ ends of Ribo-seq and 3′ ends of 3′ RACE reads were mapped to unc-54(nonstop), with read counts displayed per million uniquely mapping reads (RPM).
(B) Zoomed-in view of the boxed area in (A). Reads aligned to the poly(A) junction are shown with the sequence mapping to the 3′ UTR in black and poly(A) sequence in red. The 3′ ends of Ribo-seq reads are shown.
(C) 3′ RACE reads (top panel; 3′ ends, blue), 28–30 nt Ribo-seq reads (middle panel; 5′ ends, green), and 15–18 nt Ribo-seq reads (bottom panel; 5′ ends, orange).
(D) Zoomed-in view from (C) showing region upstream of and including the 12 rare arginines (alternating gray boxes).
(E) 5′ RACE reads mapped to unc-54(no-go). The 5′ ends of reads are displayed as fractional read abundance (STAR Methods).
(F) Multiple sequence alignment of the P loop kinase domain of NONU-1 and its homologs. See text.
