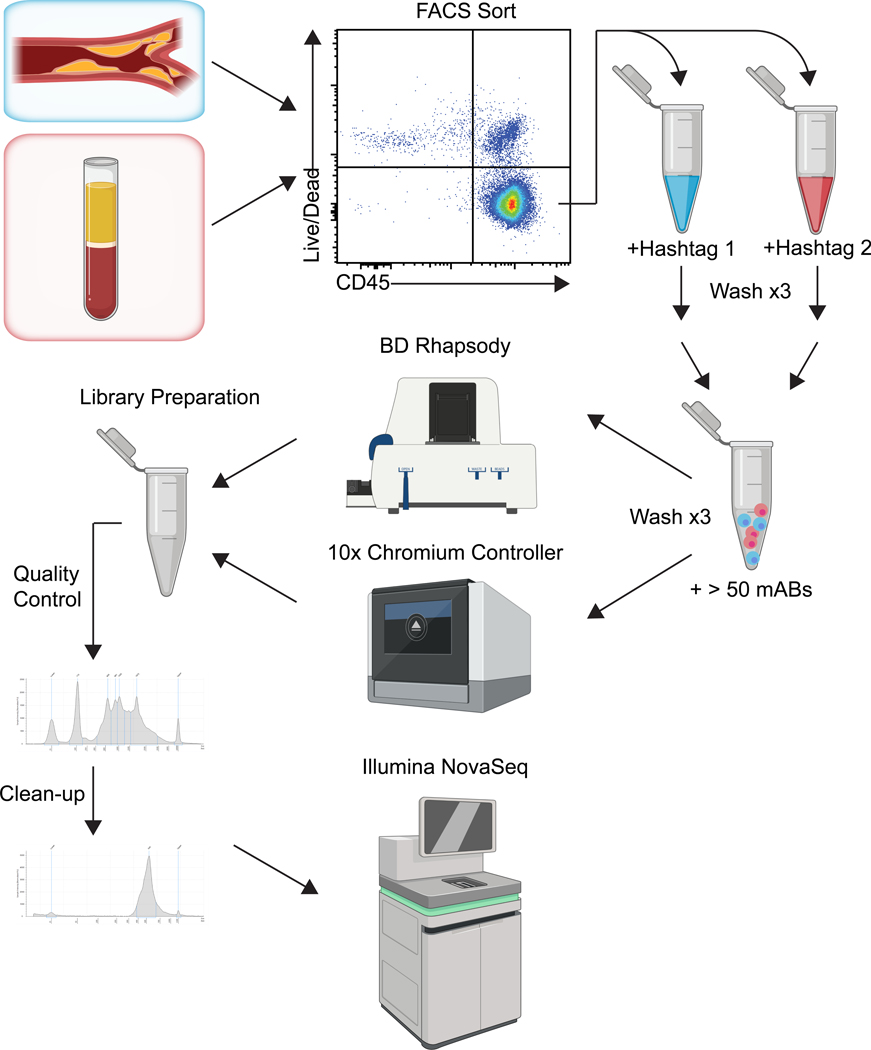
Figure 1.
Experimental workflow for CITE-seq and AB-seq experiments.
Single cell suspensions derived from artery wall (top) or PBMCs (bottom) are sorted for live cells and other desired markers (here: CD45 for leukocytes), hash-tagged, washed and incubated with the oligonucleotide-marked antibody panel (currently 50 mAbs are possible). After 3 more washes, the cells are loaded into the BD Rhapsody scanner or the 10x Chromium controller. Beads are retrieved and processed for library preparation. Pre-sequencing quality control (QC) is done by TapeStation. DNA fragments are removed, and the library is sequenced using NGS sequencers like the Illumina NovaSeq. The figure was created with BioRender.