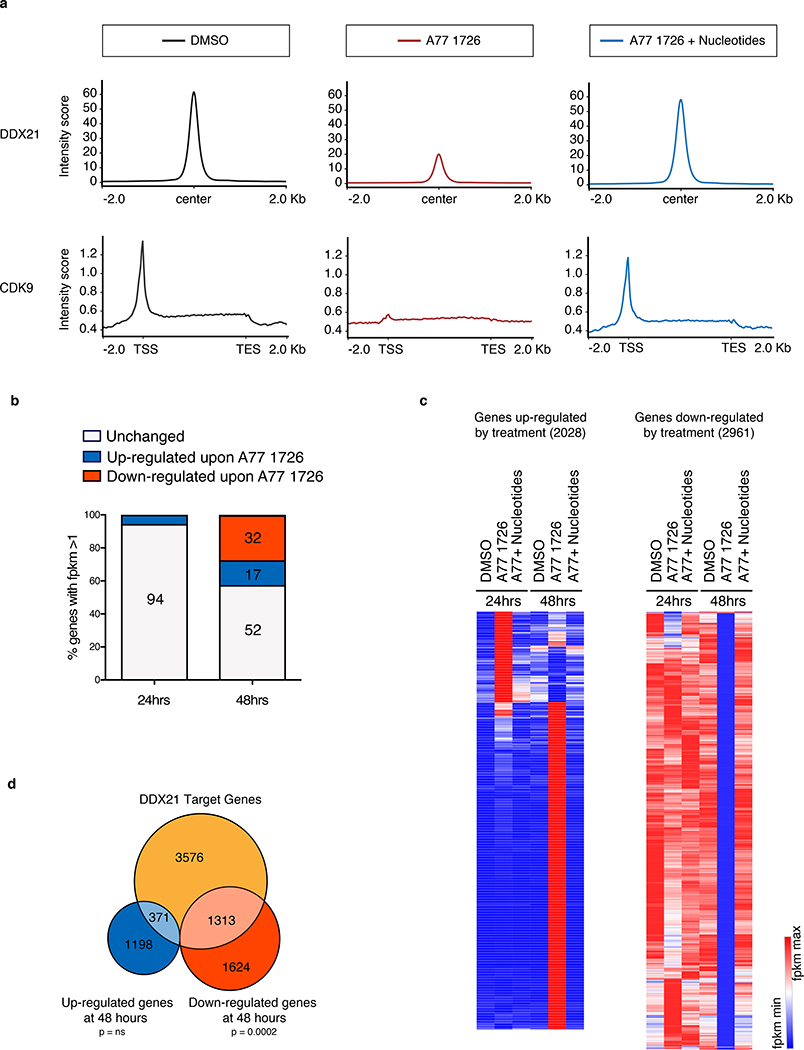
Figure 5. Ddx21 mediates transcriptional changes during nucleotide stress.
(a) ChIP-Seq for DDX21 and CDK9 in A375 melanoma cells after 24 hours of treatment. Upper panel: Metagene analysis of DDX21-bound regions. Normalized peak intensities ± 2Kb from the center of DDX21 ChIP-seq are shown. Lower panel: Metagene representation of normalized CDK9 occupancy at transcription start site (TSS), along gene bodies and at transcription end site (TES) are shown. (b) Percentage of differentially expressed genes (DEGs) in A375 melanoma cells after 24 hours and 48 hours in the presence of A77 1726. DEG criteria: FPMK ≥1 after treatment, Log2 fold change ≥1 or ≤ −1. (n = 3 biologically independent experiments). (c) Hierarchical clustering heatmap of FPKM values from genes up-regulated (2028) and down-regulated (2961) at 24 hours and 48 hours post DHODH inhibition plus or minus nucleotide supplementation. (d) Venn diagram of DDX21 target genes with reduced DDX21 binding upon A77 1726 treatment as defined by ChIP-Seq (orange); genes up-regulated by RNA-Seq (blue) and genes down-regulated by RNA-Seq (red) at 48 hours post treatment with A77 1726 in A375 melanoma cells (n = 3 biologically independent experiments, Hypergeometric Test, ns = not significant; under enrichment p-value = 2.7e-58). Source data are available online.