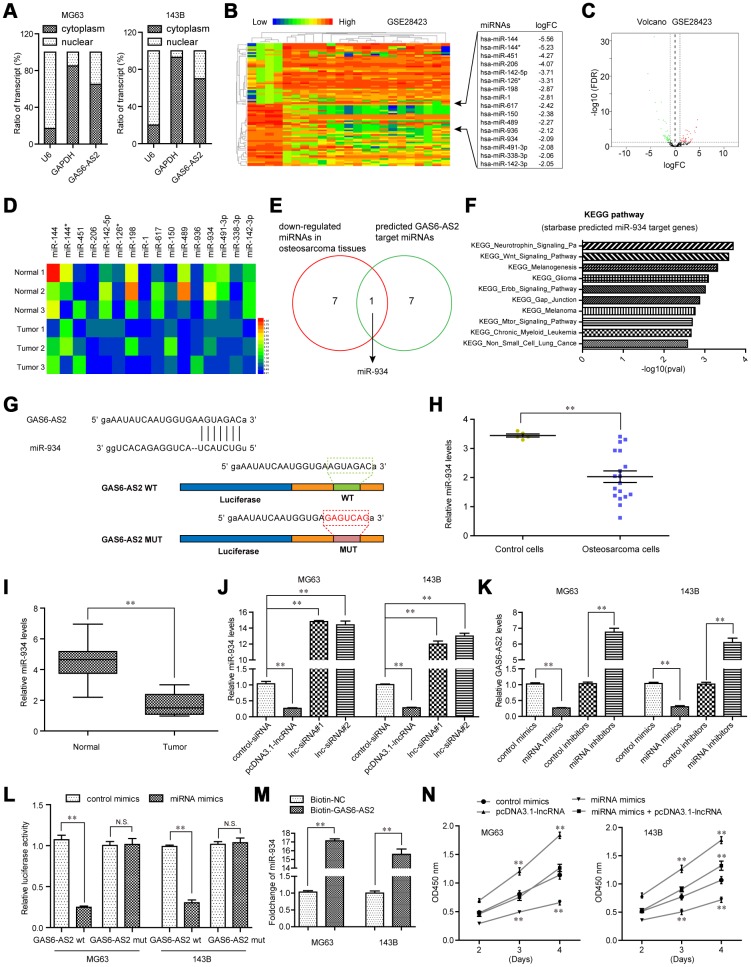
Figure 6.
GAS6-AS2 sponged miR-934 in OS cells. (A) Subcellular fractionation assays were used to determine the subcellular localization of GAS6-AS2 in OS cells. (B) Heatmap of differentially expressed miRNAs in GSE28423. (C) Volcano map. (D) Real-time PCR detected the expression of miRNAs in randomly selected three paired OS tumor specimens and normal tissues. (E) Intersection of down-regulated miRNAs in OS and “starbase” algorithm predicted GAS6-AS2 target miRNAs. (F) “starbase” algorithm analyzed the KEGG pathway of miR-934 target genes. (G) “starbase” algorithm predicted binding sites of GAS6-AS2 and miR-934. (H) Relative miR-934 expression in GSE28423 dataset. (I) Relative miR-934 expression in 157 OS samples. (J) qPCR detected miR-934 levels in MG63 and 143B cells. (K) qPCR examined the levels of GAS6-AS2. (L) Luciferase activity detection. GAS6-AS2 wt: luciferase reporter containing wild-type binding site of GAS6-AS2 and miR-934. GAS6-AS2 mut: luciferase reporter containing mutant-type binding site of GAS6-AS2 and miR-934. (M) RNA-pull down assays. (N) CCK-8 assays detected cell proliferation.