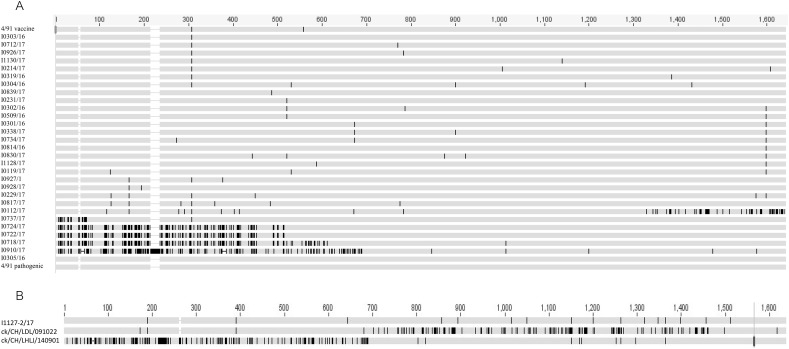
Fig. 3.
Multiple alignments of the nucleotide sequences of S1 genes performed using MAFFT version 6 (http://mafft.cbrc.jp/alignment/software/). The sequences of our 38 GI-13 viruses, 4/91 vaccine, 4/91 pathogenic strain, and six GI-13-related viruses isolated in this study were compared (A). Nine isolates, including I0813/16, I0130/17, I0213/17, I0227/17, I0232/17, I0925/17, I0522/17, I0230/17 and I0120/17, shared the same sequence with that of I0303/16 and hence, only the sequence of I0303/16 is listed in the alignment. Similarly, isolates I0320/16, I0511/16, I0339/17 and I0922/17 had the same sequences as I0301/16 and only the sequence of I0301/16 is listed in the alignment. In addition, isolates I0231/17 and I0321/16, I0305/16 and I0651/17 had the same sequences, respectively, and only I0231/16 and I0305/16 are listed, respectively, in the alignment. The deleted sequences are represented in white. The GenBank accession numbers for these genome sequences are listed in Supplemental Table 2. The sequence of isolate I1127–2/17 was compared with those of reference strains ck/CH/LHLJ/140901 and ck/CH/LDL/091022 (B).