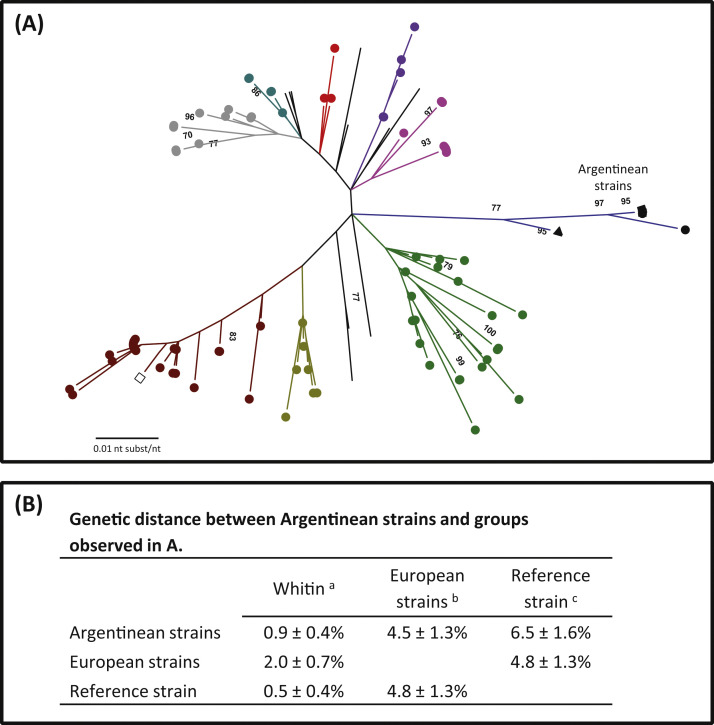
Fig. 3.
(A) Phylogenetic tree based on the nucleotide sequence of the hypervariable region (330 bp -nucleotide 1381–1710 of the Mebus strain S gene U00735.2-). Bootstrap values higher than 70% are shown. (●) Dairy strains, (■) beef strains, (▲) Argentinean reference strain Arg95, (□) Mebus reference strain. Each color represents a different geographical cluster. The evolutionary history was inferred by the Maximum Likelihood method based on the Tamura 3-parameter model. The tree was drawn to scale with branch lengths measured in the number of substitutions per site. The analysis involved 132 nucleotide sequences. Codification of each sample is shown in Fig. 1 from Supplementary material. (B) Genetic distance average between nucleotide sequences of Argentinean, European and reference strains expressed in percentage ± standard deviation.