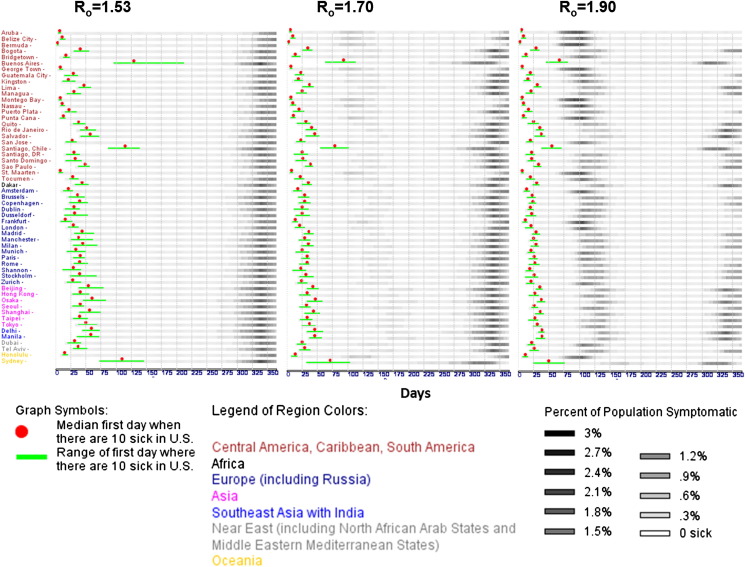
Figure 2.
Predicted disease spread time-course. Simulations were based on 100 exposed people from each international metropolitan point of origin across three reproduction numbers: 1.53, 1.7, and 1.9. The Y-axis displays origins that are first grouped by continent and then sorted alphabetically. The X-axis denotes time in days relative to the start of the disease-spread simulation. Gray-scale raster plots represent the number of infected people across time as a fraction of the aggregate U.S. metropolitan population size employed by the model. Each row shows the median disease spread for all suprathreshold trials (out of 40) in which at least 10 symptomatically infectious people appeared in the United States. In the R0 = 1.53 simulations, early disease arrival times varied from about 5 days to slightly under 225 days from disease emergence in a population. Approximately 95% of origins resulted in median early disease arrival under 75 days, with 85% of those origins resulting in median early disease arrival in less than 50 days. In the R0 = 1.7 simulations, early disease arrival time ranged from about 5 days to approximately 100 days. Approximately 95% of origins resulted in median early disease arrival in less than 50 days, with 37% of those origins resulting in median early disease arrival in less than 25 days. In the R0 = 1.9 simulations, early disease arrival time varied from about 5 days to slightly under 80 days. Approximately 96% of origins resulted in median early disease arrival under 50 days, with 53% of those origins showing median early disease arrival in less than 25 days.