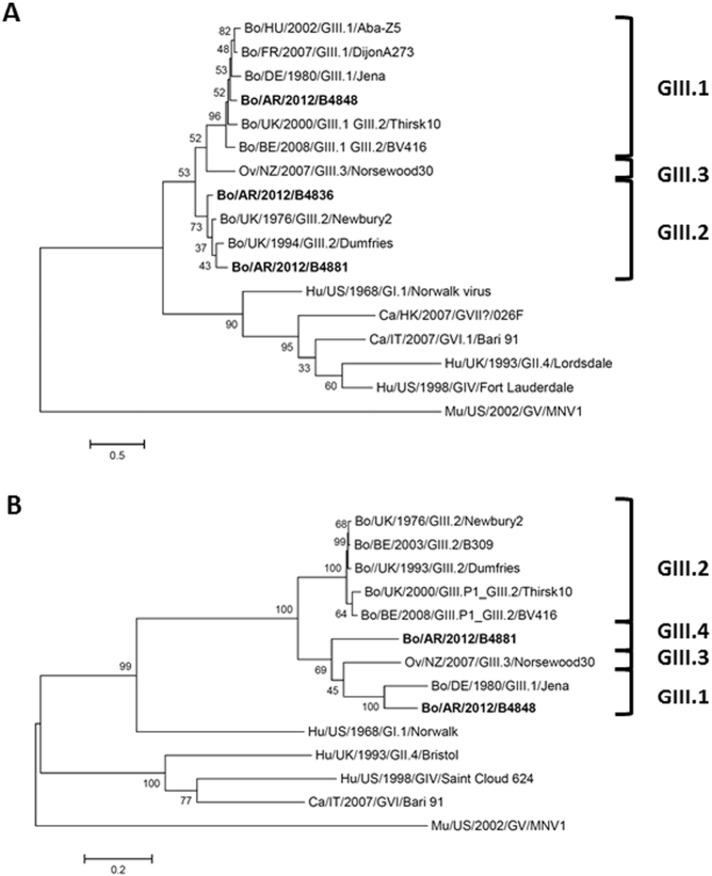
Fig. 1.
Phylogenetic trees constructed by the Maximum Likelihood method. A. Phylogenetic tree based on a partial coding region for the RNA-dependent RNA polymerase (RdRp, 532 nucleotides) of Norovirus sequences identified in this study (B4836, B4848 and B4881) and other bovine Norovirus reference sequences. The substitution model (Tamura 3-parameter + γ distribution) was selected on the basis of both lowest Akaike and Bayesian information criterion scores. The scale bar represents the phylogenetic distances expressed as units of expected nucleotide substitutions per site. B. Phylogenetic tree based on complete capsid protein (VP1) sequence of two of the bovine Norovirus sequences identified in this study (B4848 and B4881), and other bovine Norovirus reference sequences. The substitution model (LG model + γ distribution + assuming that a certain fraction of sites are evolutionarily invariable) was selected on the basis of both lowest Akaike and Bayesian information criterion scores. The scale bar represents the phylogenetic distances expressed as units of expected amino acid substitutions per site. The numbers at the nodes indicate the frequencies of occurrence for 1000 bootstrap replicate trees. The sequences detected in this study are repaired in bold case. GenBank submission numbers: 1866865 for partial RdRp sequences and 1866066 for VP1/VP2 coding sequences.