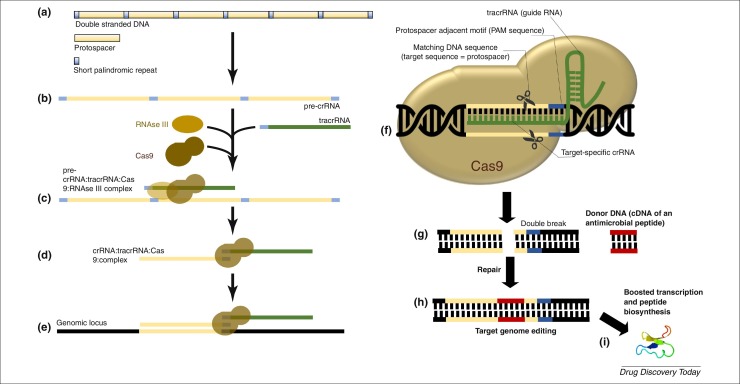
Figure 3.
CRISPR/Cas system II technology description for AMP biosynthesis mediated by genome edition. Spacer acquisition: (a) Formation of CRISPRs array by recognition and integration of foreign DNA as spacer within the CRISPR locus, or fully synthesized by genetic engineering. The protospacers are non-coding region inserted in the bacterial DNA with 24–48 bp. Adjacent to each protospacer are found 3–5 bp short DNA sequences termed protospacer adjacent motifs (PAM) crRNA processing: (b) The CRISPR array is transcribed as a long RNA (pre-crRNA) that is cleaved into crRNAs with the help of Cas proteins. An extra small RNA (tracrRNA) complementary to the repeat sequence is also synthesized. (c) The tracrRNA pairs with the repeated region of crRNA and helps in the processing of pre-crRNA into crRNA with the help of RNAse III for cleavage. Interference stage: crRNAS binds to Ca proteins (d) to form a complex that recognizes foreign DNA (e). A single multifunctional protein, Cas9, recruits crRNA and tracrRNA to cleave the recognized foreign DNA using internal endonuclease domains (f). All the process is based in the recognition and pairing of the PAM and the foreign DNA. Double strand breaks (DSBs) are generated (g). A donor DNA containing the coding sequence of an AMP of interest is integrated in the site of the DSBs by homologous recombination (i), and after gene expression, the AMP is extracted and purified (h).