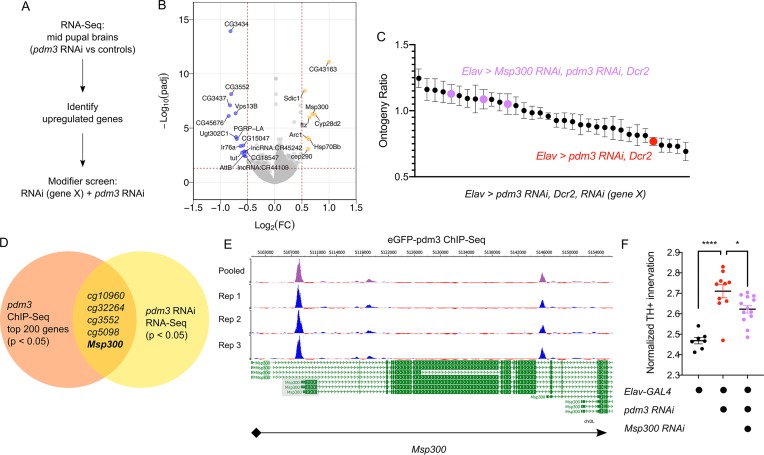
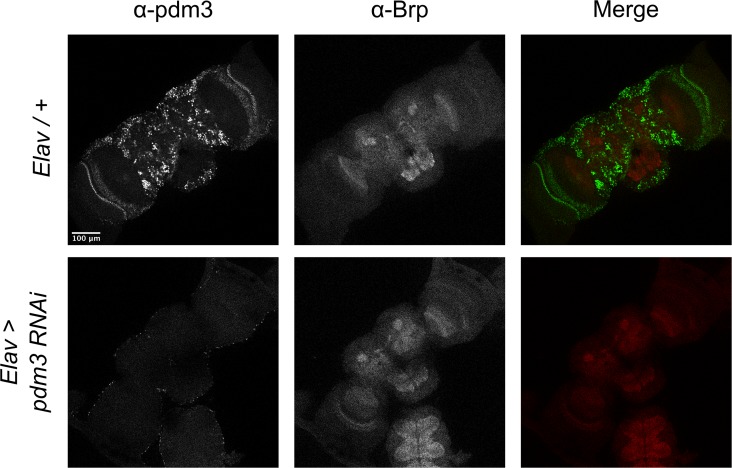
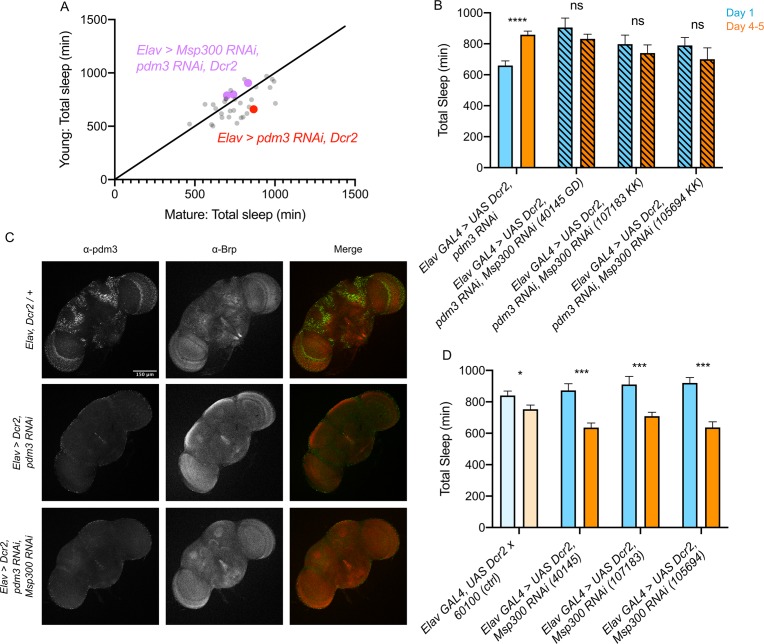
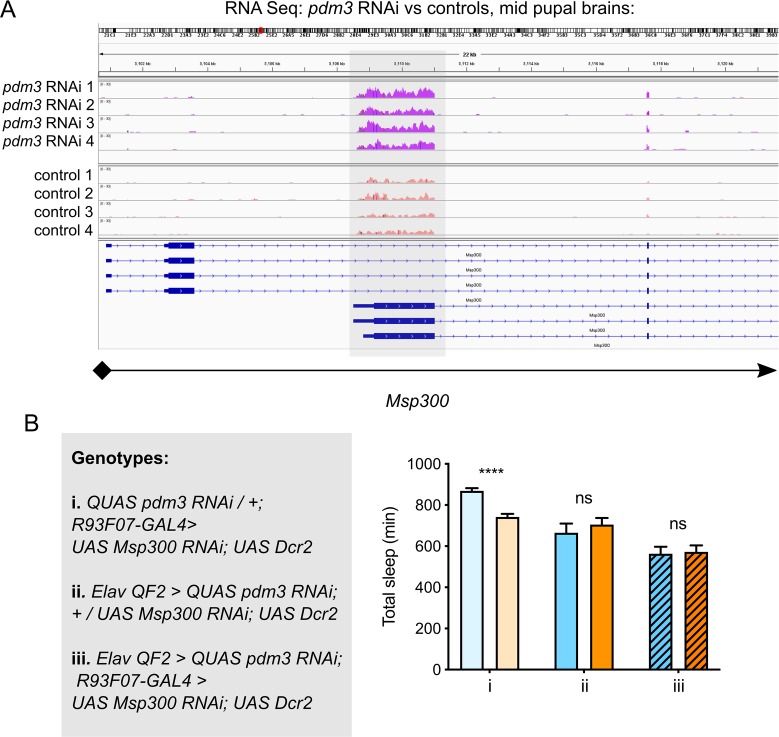
Figure 8. Pdm3 controls expression of the synaptic gene Msp300 to regulate sleep ontogeny.
(A) Scheme of RNA-Seq and subsequent modifier screen. (B) Volcano plot of RNA-Seq data highlighting significant changes in gene expression with pdm3 knockdown compared to controls in mid-pupal brains. Labeled genes have −log10(padj) >1.3 and absolute value of log2(Fold Change)>0.5. Yellow = increased expression, blue = decreased expression upon pdm3 knockdown (n = 4 biological replicates per genotype, 40 brains per replicate for RNA-Seq). (C) Modifier screen with co-expression of RNAi targeting upregulated genes from RNA-Seq alongside pdm3 RNAi (n ≥ 16 per genotype/age). (D) Overlap of hits from RNA-Seq and pdm3 ENCODE ChIP-Seq experiments. (E) Control-normalized peaks of PDM3 binding within the Msp300 gene. The strongest binding peak occurs upstream of the first exon in the RD, RL and RB transcript isoforms (shaded gray box). (F) Quantification of TH+ staining in the dFSB with Msp300 RNAi and pdm3 RNAi (n = 7, 10, 13 left to right). RNA-Seq statistical analysis is detailed in Materials and methods. ****p<0.0001, *p<0.05; ANOVA with Tukey’s test (F).