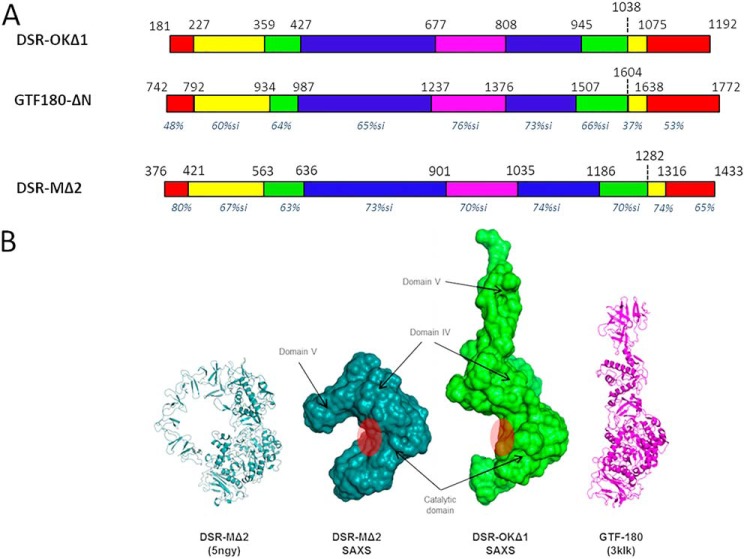
Figure 2.
A, schematic structural organization of DSR-OKΔ1 compared with GTF-180 and DSR-MΔ2, based on amino acid alignment with both enzyme primary structures and structural analyses. Nonaligned zones are not represented. Domain V is represented in red, domain IV in yellow, domain A in blue, domain B in green, and domain C in purple. For each domain, the percentage of similarity with that of DSR-OK is shown in italics (pairwise alignments with BlastP tool). B, SAXS reconstruction of DSR-OKΔ1 compared with DSR-MΔ2. From left to right: DSR-MΔ2 crystal structure, SAXS ab initio envelope of DSR-MΔ2, SAXS ab initio envelope of DSR-OKΔ1, and GTF-180 crystal structure. Assignment of domains is based on superposition between the DSR-MΔ2 crystal structure and SAXS envelopes. The approximate position of the catalytic cleft is shown with a red circle.