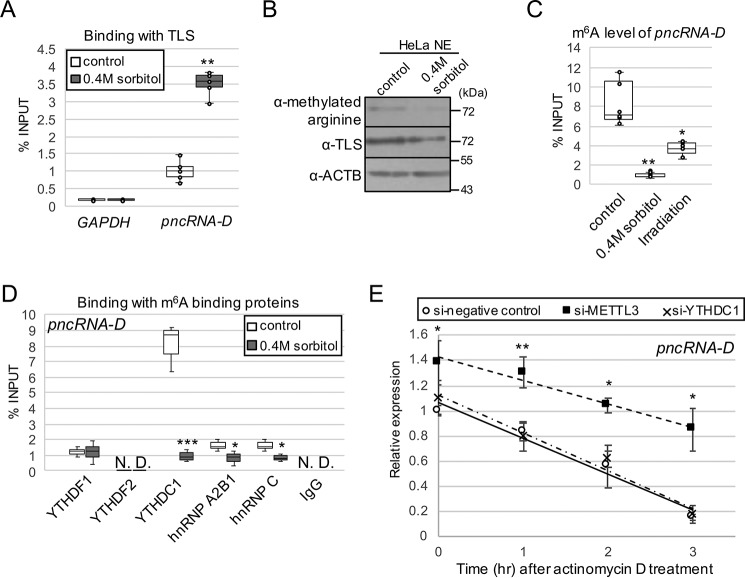
Figure 3.
pncRNA-D is highly m6A-modified, and the modification destabilizes pncRNA-D. A, the interaction between pncRNA-D and TLS was detected by RIP assay using TLS antibody with HeLa cells before and after osmotic stress (n = 5). B, Western blotting analysis using HeLa NE with or without 0.4 m sorbitol treatment. The arginine-methylated TLS level was compared with the total TLS level. ACTB was used as a loading control. A representative image of three independent experiments is shown. C, RIP assay with HeLa cell after osmotic stress or irradiation by m6A antibody (n = 5). D, RIP assay as in A with indicated antibodies of m6A recognition proteins. pncRNA-D bound to each recognition protein was measured by RT-qPCR (n = 3). E, relative expression level (normalized to GAPDH) of pncRNA-D after actinomycin D treatment with HeLa cells treated with siRNAs of negative control, METTL3, or YTHDC1. Expression levels at 1, 2, and 3 h after actinomycin D treatment are shown. Expression levels at 0 h in control cells were set to a value of 1.0 (n = 5). *, p < 0.05; **, p < 0.01; ***, p < 0.005.