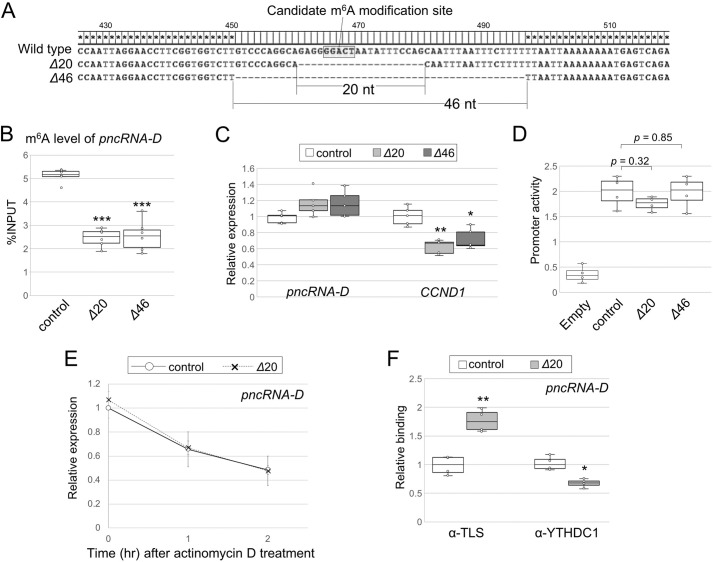
Figure 5.
Deletion of GGACU motif in pncRNA-D decreases m6A modification level and alters interaction with RNA-binding proteins, but not RNA stability. A, sequences deleted by CRISPR/Cas9 are described. HAP1 clones named Δ20 and Δ46 lack 20 and 46 nucleotides around GGACT, respectively. B, RIP assay with WT or GGACT deleted HAP1 cells by anti-m6A antibody (n = 5). C, relative expression level (normalized to GAPDH) of pncRNA-D and CCND1 of WT or GGACT-deleted HAP1. Expression levels in control cells were set to a value of 1.0 (n = 5). D, luciferase reporter assay with control or GGACT deleted promoter region of CCND1 (n = 4). E, relative expression level (normalized to GAPDH) of pncRNA-D after actinomycin D treatment in WT or GGACT deleted HAP1. Expression levels of 1 and 2 h after actinomycin D treatment are shown. Expression levels at 0 h in control cells were set to a value of 1.0. F, RIP assay as in B with antibodies against TLS or YTHDC1. pncRNA-D bound to each protein was measured by RT-qPCR (n = 4). *, p < 0.05; **, p < 0.01; ***, p < 0.005 in all the figures.