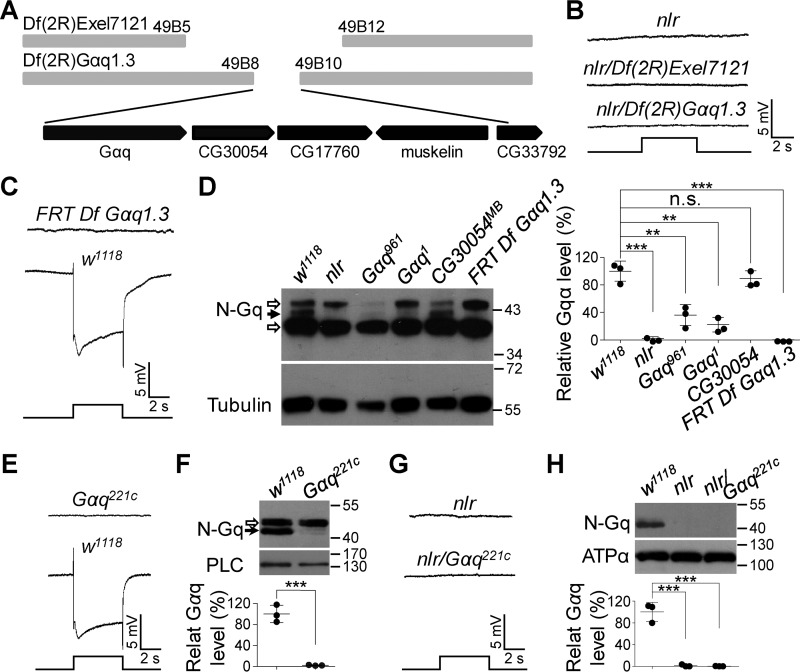
Figure 2.
Gαq gene mutations cause the abolished light responses in nlr mutants. A, annotated deletion region of the deficient chromosome Df(2R)Exel7121 and Df(2R)Gαq1.3. Annotated genes covered by Df(2R)Gαq1.3 are presented in the bottom panel. B, nlr over either Df(2R)Exel7121 (nlr/Df(2R)Exel7121) or Df(2R)Gαq1.3 (nlr/Df(2R)Gαq1.3) show no photoresponses. Event markers represent 5-s light pulses, and scale bars are 5 mV. C, ERG response of the clones of Df(2R)Gαq1.3-covered gene nulls in the retina. D, Western blots of Gαq protein levels in the various mutants. Each lane was loaded with half of the isolated retina. Gαq961 and Gαq1 mutants were used as negative controls, whereas CG30054MB mutants were used as positive controls. The specific Gαq band and nonspecific bands are marked with a black arrow and open arrows, respectively. Quantification of Gαq levels for each genotype is presented in the right panel. E, ERG response of Gαq221c null mutant clones in the retina. Event markers represent 5-s light pulses, and scale bars are 5 mV. F, Western blots of Gαq protein levels in w1118 and Gαq221c null mutant photoreceptor cells. The specific Gαq band and nonspecific bands are marked with a black arrow and an open arrow, respectively. The eye-specific protein PLC was used as a loading control. Quantification of Gαq levels for each genotype is presented in the lower panel. G, nlr/Gαq221c flies show no photoresponses. Event markers represent 5-s light pulses, and scale bars are 5 mV. H, Western blots of Gαq protein levels in the various mutants. Each lane was loaded with half of the isolated retina. Quantification of Gαq levels for each genotype is presented in the lower panel.