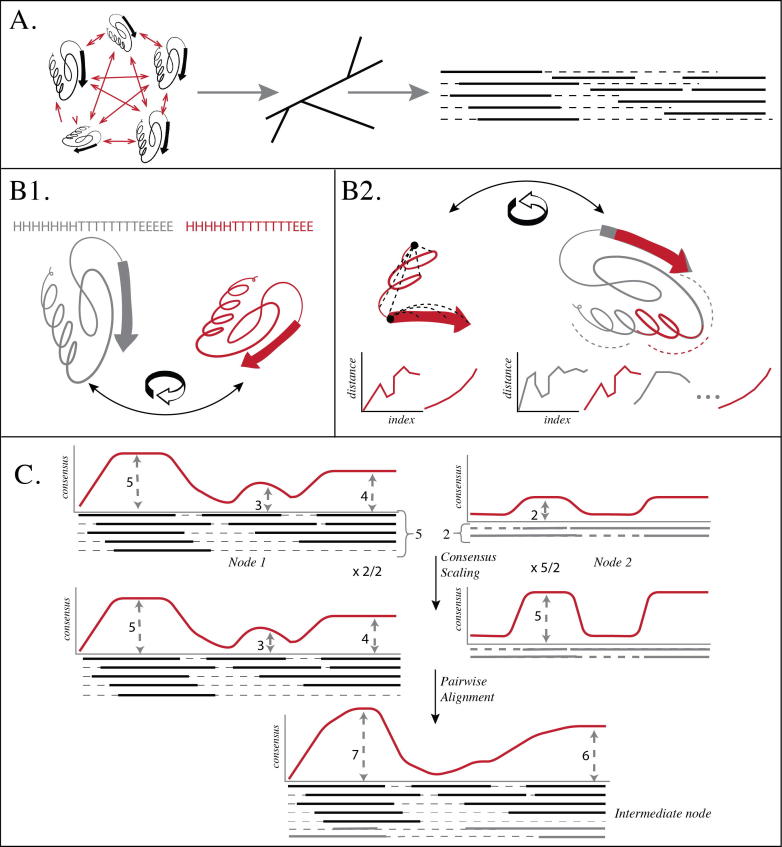
Fig. 1.
A. Caretta’s multiple structure alignment workflow: an all vs. all pairwise alignment step, followed by construction of a guide tree and progressive alignment. B. The two approaches for initial rotation and superposition of two structures used in pairwise alignment: 1) aligning secondary structure codes and 2) dynamic time warping on one-dimensional signals of distances from all residues to the first or last residue in a segment. C. The guide tree specifies the two neighbors to combine at each progressive alignment step. These two neighbors can either be protein structures or previously combined intermediate nodes. A new intermediate node is created at each step by aligning and combining the two neighbors. The alignment step takes into account the number of times each position has been aligned in each of the two neighbors weighted by the number of structures in each neighbor (consensus row, shown in red). This ensures that when the difference of the two is taken in calculating (Eq. (3)), positions with fewer gaps get higher scores. After alignment, the consensus row of the new intermediate node keeps track of the number of residues aligned at each position.