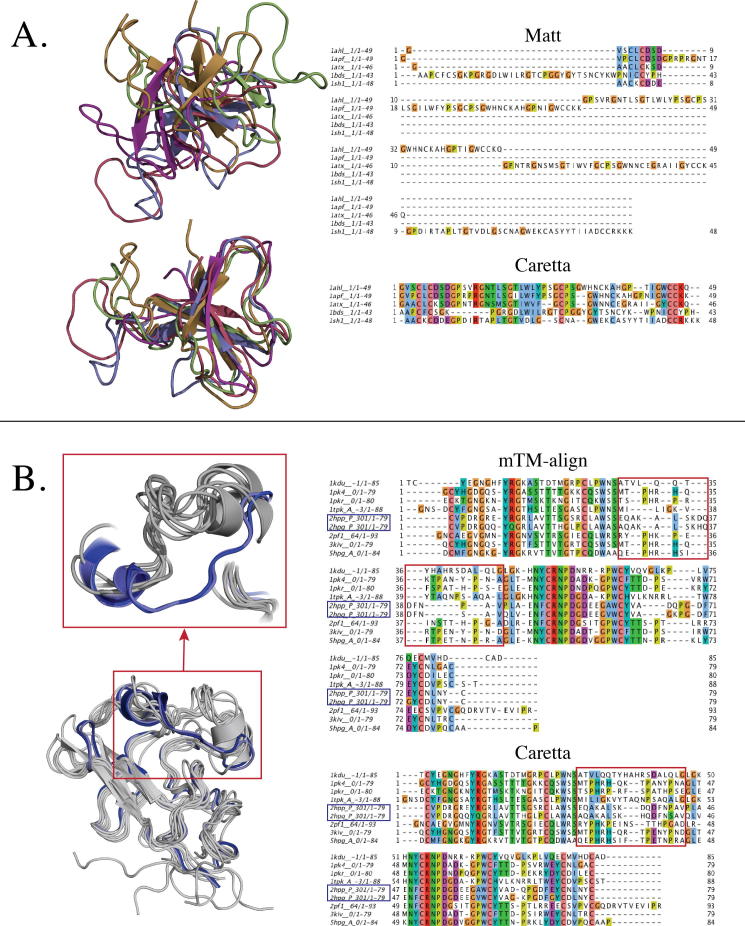
Fig. 3.
Two examples of protein families in which Caretta finds a better alignment than Matt and mTM-align. A. structures from the “seatoxin” family (a collection of toxins released by sea anemones), superposed according to alignments made by Matt (top) and Caretta (bottom) respectively, with the alignments shown on the right. B. structures from the “Kringle” family (PFAM ID: PF00051) superposed according to Caretta’s alignment. Two structurally divergent proteins are highlighted in blue and the region of divergence is highlighted in red, both in the structure superposition and in the corresponding alignments on the right. These two groups of proteins are obtained from the Homstrad benchmark dataset [26].