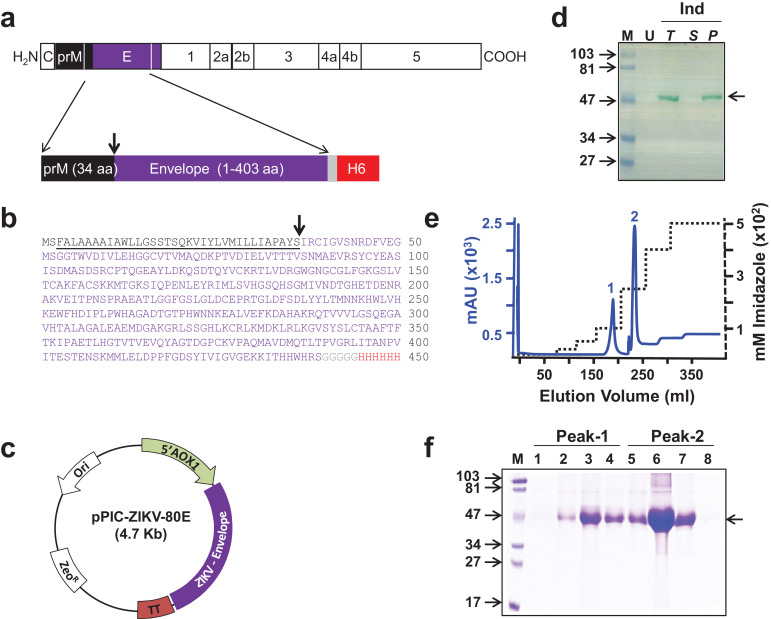
Fig. 1.
Design, expression, and purification of ZIKV-80E antigen. (a) Schematic representation of the ZIKV polyprotein. Proteins prM and E are indicated by black and purple boxes, respectively. The region of the polyprotein included in the antigen design is bounded by the two white lines in the C-terminal regions of prM and E. Shown below is the schematic representation of the design of the recombinant ZIKV-80E antigen consisting of the last 34 aa residues of prM (black box) and the first 403 aa residues of E (purple box). The C-terminally located grey and red boxes denote the pentaglycyl peptide linker and the hexa-histidine (H6) tag, respectively. (b) Predicted aa sequence of the recombinant ZIKV-80E antigen. The colour scheme corresponds to that shown in ‘a’. prM aa residues are underlined. The N-terminal dipeptide ‘MS’ was introduced during cloning. The downward arrow in panels ‘a’ and ‘b’ denotes the signal peptide cleavage site. (c) Map of the ZIKV-80E expression plasmid, pPIC-ZIKV-80E. The ZIKV-80E gene (ZIKV-Envelope) is inserted between the AOX1 promoter (5′ AOX1) on the 5′ side and the transcriptional terminator (TT) on the 3′ side. The vector carries the zeocin selection marker (ZeoR), which is functional in both E. coli and P. pastoris, and the E. coli origin of replication (Ori), for bacterial propagation. (d) Analysis of localisation of recombinant ZIKV-80E protein in P. pastoris. An aliquot of methanol-induced (Ind) culture of P. pastoris was lysed with glass beads and separated into supernatant (S) and pellet (P) fractions. Total (T) extract prepared from an equivalent aliquot of the induced culture, S, and urea-solubilised P fractions were run on SDS-polyacrylamide gel and subjected to Western blot analysis using mAb 24A12. T extract prepared from an equivalent aliquot of the un-induced (U) culture was analysed in parallel. Pre-stained protein markers were analysed in lane ‘M’. Their sizes (in KDa) are indicated to the left. The arrow on the right indicates the position of the recombinant ZIKV-80E protein. (e) Chromatographic profile of recombinant ZIKV-80E purification by Ni2+ affinity chromatography, starting from P fraction of induced cell lysate under denaturing conditions. The solid blue and the dotted black curves represent the profiles of UV absorbance (at 280 nm) and the imidazole step-gradient, respectively. Bound protein was eluted as two peaks (1 and 2). (f) Coomassie-stained SDS-polyacrylamide gel analysis of fractions corresponding to peaks 1 (lanes 1–4) and 2 (lanes 5–8) shown in panel ‘e’. Protein markers were analysed in lane ‘M’. Their sizes (in KDa) are indicated to the left. The arrow on the right indicates the position of the recombinant ZIKV E protein.