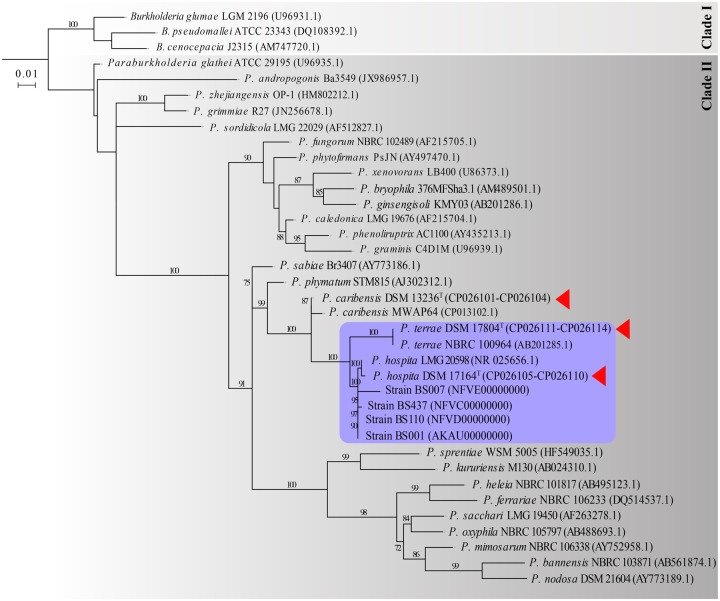
Fig. 1.
—Maximum-likelihood tree based on the V2–V6 (>1,000 bp) regions of the 16S rRNA gene. Accession numbers for the sequences used for each organism are provided in brackets after the organism’s name. The tree was constructed using RAxML (nucleotide substitution model GTRCAT), default algorithm settings (hill-climbing), with bootstrap value of 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box: proposed “species cluster.” Red triangles: type strains.