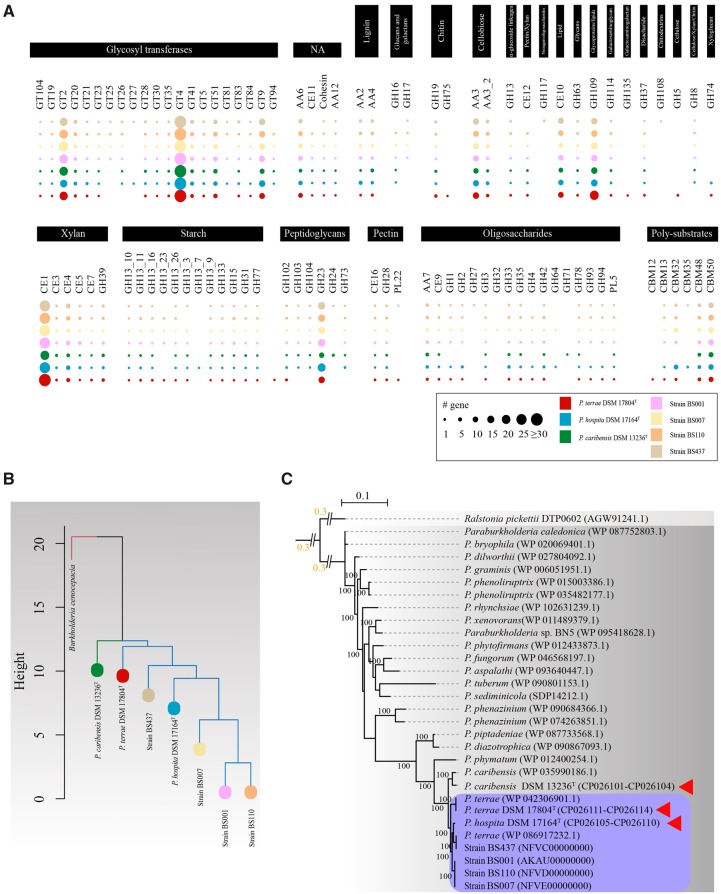
Fig. 6.
—(A) Gene number profile of predicted CAZYmes family proteins in Paraburkholderia terrae DSM 17804T, Paraburkholderia hospita DSM 17164T, and Paraburkholderia caribensis DSM 13236T, next to fungiphilic strains BS001, BS007, BS110, and BS437. Color code based on the strain is indicated. Number of genes is indicated by the dot size. (B) Hierarchical cluster analysis based on CAZyme gene number for P. terrae DSM 17804T, P. hospita DSM 17164T, P. caribensis DSM 13236T, strains BS001, BS007, BS110, and BS437. B. cenocapacia J2315 was used as an outgroup. (C) Phylogenetic analysis of GH3 type proteins across all strains, and the top hits of PSI-BLASTP. The tree was built with RAxML using amino acid substitution model PROTGAMMA, default matrix setting (Dayhoff) and hill-climbing algorithm, with bootstrap value of 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box represents the proposed “species-cluster,” and red triangles indicate the type strains.