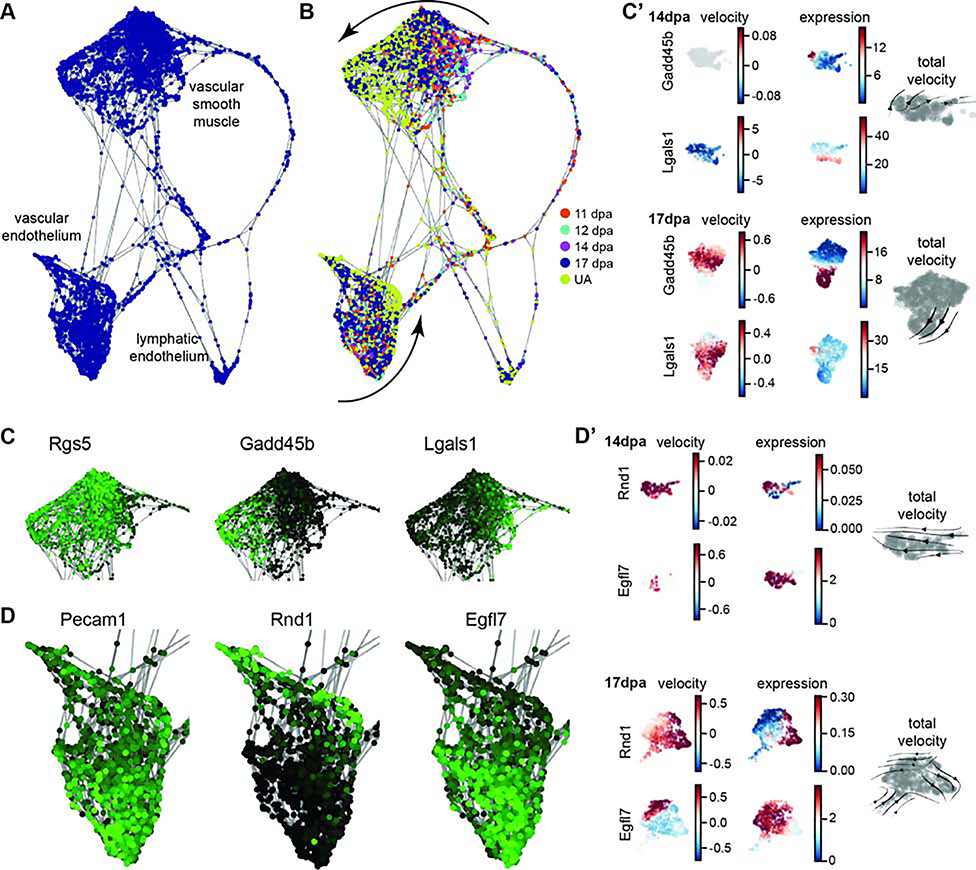
Figure 4. Vasculature differentiation trajectory of integrated data set.
Computationally defined SPRING trajectory analysis of cells from the integrated data set vascular clusters 6, 9, and 18. (A) Force-directed plot of cells showing clusters of VSMs, vascular endothelial cells, and lymphatic endothelial cells. (B) SPRING plot as in (A) with regenerative stages of each cell colored coded: 11dpa (orange), 12dpa (light blue), 14dpa (purple), 17dpa (dark blue), unamputated (yellow). Differential clustering of blastema cells and unamputated cells suggests tissue specific differentiation of VSMs and the vascular endothelium (curved arrows). (C) Gene expression overlay on VSMs. Rgs5 is expressed in all cells, Gadd45b is more highly expressed in UA cells, and Lgals1 is more highly expressed in blastema cells. High expression is in green and low expression is black. (C’) UMAP plots of RNA velocity and expression data for individual genes Gadd45b and Lgals1 at 14dpa and 17dpa; total RNA velocity stream plot for each stage in gray (right). (D) Gene expression overlay on vascular endothelial cells. Pecam1 is expressed in all cells, Rnd1 is more highly expressed in UA cells, and Egfl7 is more highly expressed in blastema cells. (D’) UMAP plots of RNA velocity and expression data for individual genes Rnd1 and Egfl7 at 14dpa and 17dpa; total RNA velocity stream plot for each stage in gray (right).