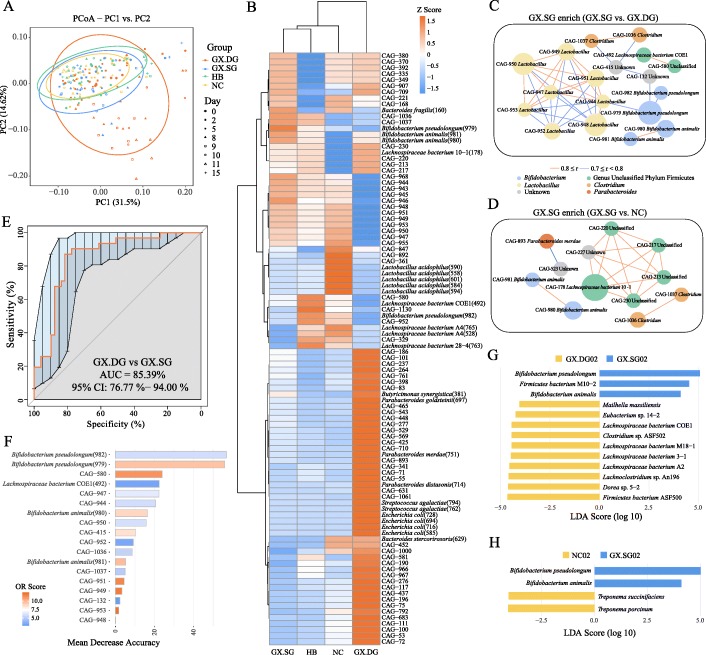
Fig. 3.
The survivability of infected mice is closely associated with the enrichment of several bacterial species. Experimental description references to Fig. 2. Of which, 177 representative samples from all groups collected on days 0, 2, 5, 8, 9, 10, 11, and 15 post-infection (57 samples from the GX.DG group and 40 samples each from the GX.SG, NC, and HB groups) were selected for metagenomic sequencing analyses. a PCoA of the weighted UniFrac distances among the GX.DG, GX.SG, HB, and NC groups. b Heat map of the relative abundances of CAGs. c, d Co-occurrence networks generated from the comparisons of the GX.SG group with the GX.DG (c) and NC (d) groups. The co-occurrence network inferred for each fecal sample according to Spearman’s correlations was stratified by the infection type. Each node in the network indicates a CAG, and the node size represents the average relative abundance of one CAG. Only the CAGs enriched in the GX.SG group compared with the GX.DG or NC group (OR > 2) were retained. e ROC curve of the final random forest model constructed using the relative abundances of 18 CAGs enriched in the GX.SG group compared with the GX.DG group. The AUC statistic is a summary measure of classifier performance. AUC values close to 1 indicate that a high true positive rate was achieved with a low false-positive rate (ideal performance), whereas AUC values close to 0.5 indicate random performance. f Variable importance according to the mean decrease in accuracy of the random forest classifier. The mean decrease in accuracy obtained after the permutation of a variable shows the importance of the variable with respect to its contribution to the accuracy of the random forest classifier. Variables with a high mean decrease in accuracy contribute more to the classification of the data than variables with a low mean decrease in accuracy. g, h LEfSe comparison of gut microbes at the species level between the GX.SG and GX.DG groups (g) and the GX.SG and NC groups (h) on day 2 post-infection