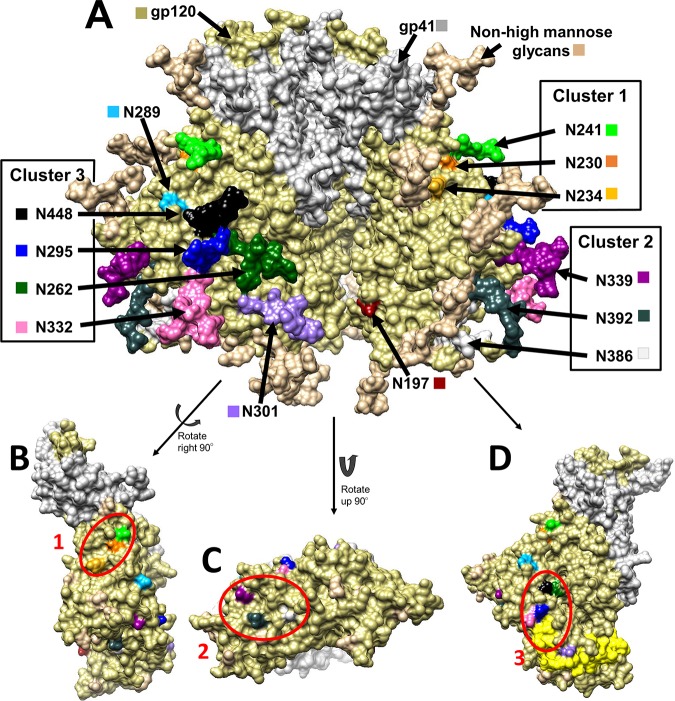
FIG 1.
(A) Structure of the HIV gp160 trimer in the unbound, closed state, i.e., in a state not bound to CD4 or CCR5. This structure of clade B JR-FL includes glycosylation (PDB accession number 5FYK) (118) and was prepared using the UCSF Chimera program (119, 120). gp120 is shown in khaki, and gp41 is shown in white. (B to D) gp120 monomers with glycosylation hidden to show the underlying amino acids that fall into clusters. Glycosylation sites (Asn) are shown in a variety of colors, as labeled in the diagram. (B) Cluster 1 encompasses the glycans at N230 (orange), N234 (yellow), and N241 (green). (C) Cluster 2 encompasses the glycans at N339 (dark magenta), N386 (dim gray), and N392 (dark slate gray). (D) Cluster 3 encompasses the glycans at N295 (blue), N332 (hot pink), N262 (dark green), and N448 (black). The V3 loop is highlighted in yellow to demonstrate the proximity of the glycans at N295 and N332.