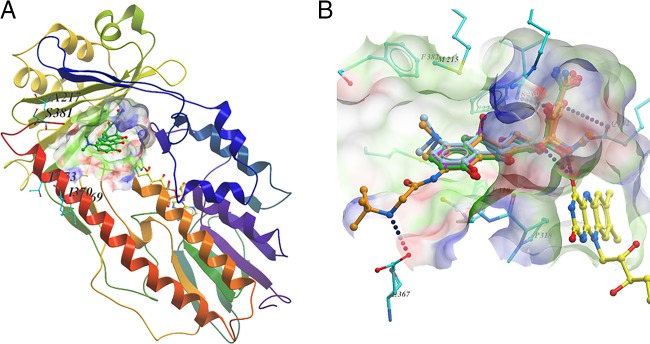
FIG 1.
Homology modeling and molecular docking of Tet(X5). (A) Cartoon representation of the modelled Tet(X5) structure in complex with ligand (carbon in green) and FAD (carbon in yellow). Nonconserved residues within 10 Å of the ligand are shown as sticks. A surface representation of the substrate-binding site is illustrated. (B) Predicted binding conformation of four tetracyclines at the substrate-binding site of the modelled Tet(X5) structure. The binding conformations suggest that the tetracycline core has essentially the same interactions with Tet(X5). Hydrogen bonds are depicted by blue dotted lines.