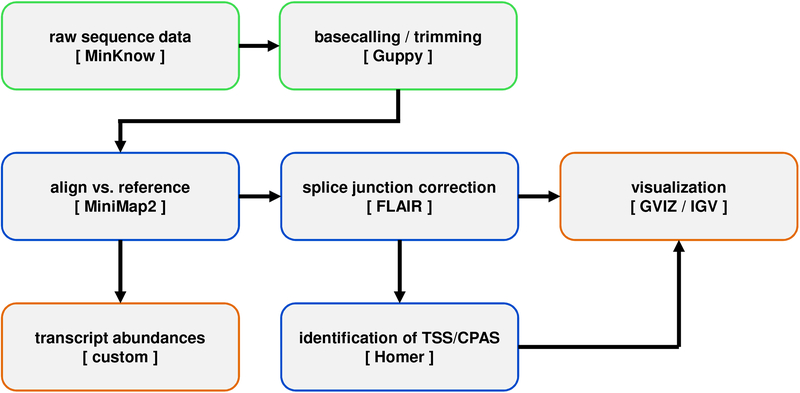
Figure 3. Computational workflow for rapid alignment and analysis of viral dRNA-Seq data.
Following the acquisition of raw sequence data produced by MinKNOW during a nanopore run, Guppy is used to rebasecall data, trim the adapter sequence, reverse the orientation of the sequence read (from 3’->5’ to 5’->3’), and replace uracil bases with thymine bases. Processed sequence reads are aligned against a chosen reference genome sequence using MiniMap2 and (optionally) Illumina data are used in conjunction with FLAIR to perform splice-site correction. At this stage, aligned datasets may be visualized using standard tools such as IGV or GVIZ before subsequent processing to produce transcript abundance counts (using custom scripts) and/or to identify TSS and CPAS (using Homer).