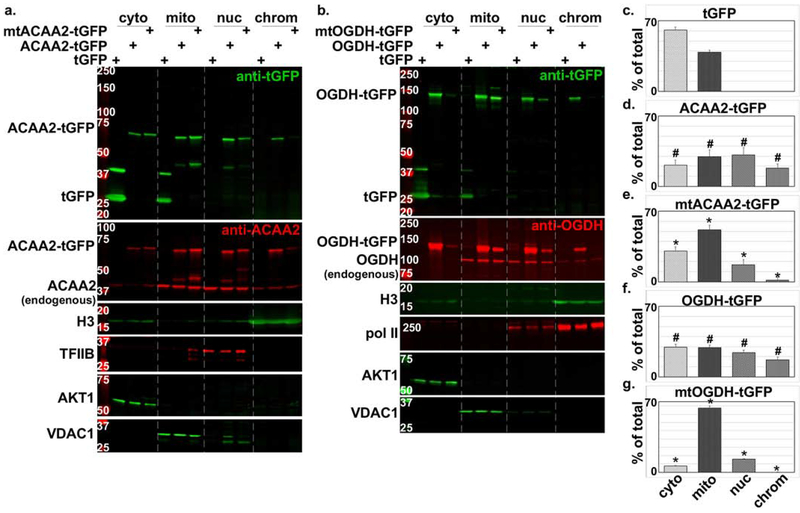
Figure 3. Nuclear localization of ACAA2 and OGDH confirmed by tGFP-fusion proteins and NLS mutations.
a-b. Cardiac myocytes were infected with a 10-20 moi of recombinant adenoviruses harboring turbo-GFP (tGFP) or a. wt ACAA-tGFP or an NLS mutant (mtACAA2-tGFP), or b. wt OGDH or an NLS mutant (mtOGDH-tGFP). After 18 h, the cellular proteins were fractionated into cytosol (cyto), mitochondrial and membrane (mito), nucleoplasm (nuc), and chromatin-bound (chrom) protein fractions that were then analyzed by Western blotting for the proteins listed on the left of each panel. The fusion proteins were detected by anti-GFP (upper panels, a-b) and anti-ACAA2 or anti-OGDH (second from the top panels, a-b), which also detect the endogenous proteins. AKT1, VDAC1, TFIIB or Pol II, and H3, were immunodetected for use as internal controls for the corresponding cell fractions. The signals for the c. tGFP, and tGFP-fusion proteins d. ACAA2-tGFP, e. mtACAA2-tGFP, f. OGDH-tGFP, g. mtOGDH-tGFP, were quantified using imageJ, normalized to internal controls, and plotted as the mean ± SEM of % total protein in all 4 fractions. Error bars represent SEM, N=3 from 3 repeats. #p ≤ 0.05 vs. tGFP, *p = 0.0095 vs. wt tGFP-ACAA2 or -OGDH fusions, in corresponding fractions.