Fig. 6.
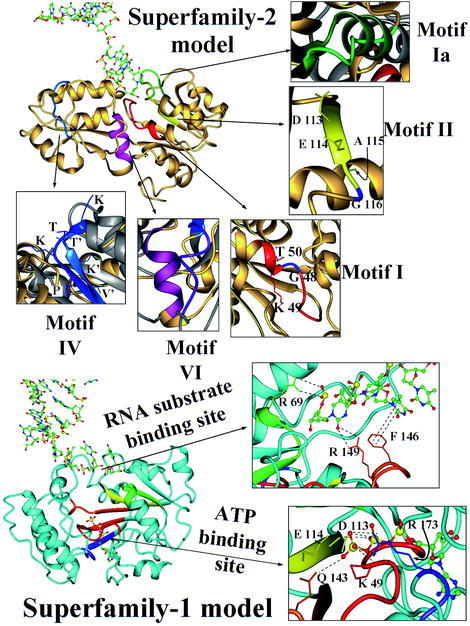
General view of the SF2 and SF1 models. Ribbon plots of the SF2 model (at the top) and SF1 model (at the bottom). Helicase motifs of helicase SF1s and SF2s are identified by colors, while the original crystallographic structure of MjDEAD (with PDB code 1hv8) is in grey and the models are in gold (for SF2 model) and cyan (for SF1 model). A detailed view of the motifs of SF2 model is shown at the top of the figure: motif I is in red (plus a conserved Gly in blue), motif Ia in green, motif II in yellow (showing a different DEAG, instead of DEAD/DEAH signature and indicating the Gly residue in blue), motif IV is in blue and motif VI in magenta. The motifs of SF1 model are indicated in red (motif I), green (motif Ia), yellow (motif II), orange (motif III) and blue (motif IV), and a detailed view of the ATP binding site implicating motif I (Lys 49), motif II (Asn 113, Glu 114), motif III (Gln 143), and motif IV (Arg173) is indicated at the margin (sequence numbering according to the model, see text). A detailed view of the DNA binding is also indicated, showing the interactions of motif Ia (Arg 69) and motif III (Arg 149 and Phe 146) with the nucleotide chain.
