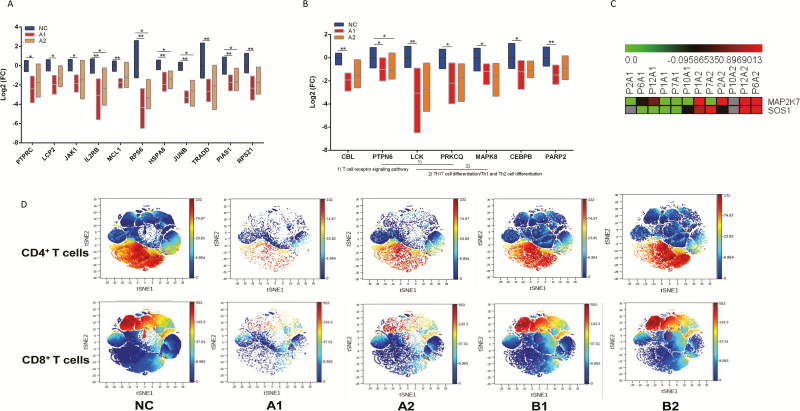
Figure 4.
A, FCs of 11 of 14 genes from Figure 2D that were compared between the A1 and A2 groups using Kruskal-Wallis test (all P < .05). Statistical significance was set at a 2-sided P value < .05. B, The FCs of group A–specific DEGs including CBL, PTPN6, LCK, PRKCQ, CEBPB, MAPK8, and PARP2 were compared among NC, A1, and A2 group using Kruskal-Wallis test (all P < .05). Statistical significance was set at a 2-sided P value < .05. C, The DEG expression profiles of mRNAs are shown by a heatmap, and the colors represent the FC values. DEGs of group A2 relative to A1 are shown by a heatmap, including MAP2K7 and SOS1, which were upregulated in group A2 after initial treatment. D, The Mass Cytometry (CyTOF)-based analysis discovered T-cell signatures in peripheral blood of patients with COVID-19. Expression patterns of T cells, CD4+ T cells, and CD8+ T cells in PhenoGraph analysis. Populations identified by the relative expression of CyTOF markers are indicated by differences in expression patterns in the NC, A1, A2, B1, and B2 groups. *P < .05, **P < .01. Abbreviations: COVID-19, coronavirus disease 2019; DEG, differentially expressed gene; FC, fold-change; Th, T-helper.