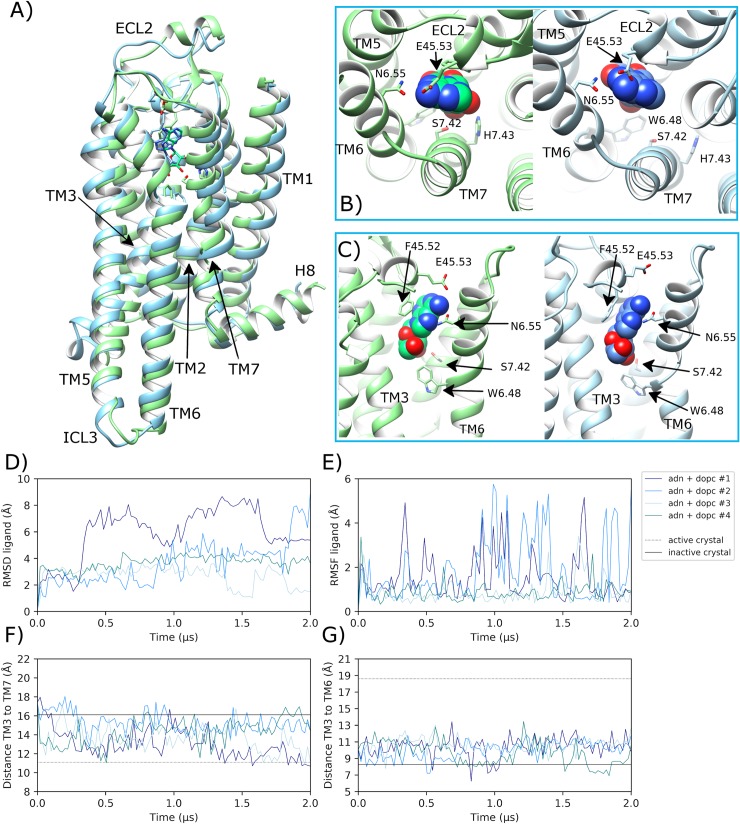
Fig 4. Transition towards an intermediate conformation of adenosine-bound A2aR in DOPC membrane.
A) Superposition of the intermediate state crystal structure of A2aR (PDB entry: 2YDO, light green) and an MD-generated conformation achieved within a DOPC membrane (blue, belonging to replica #3 from 1.5 μs) bound to adenosine, showing B) and C) protein-agonist interactions in the orthosteric pocket with adenosine atoms displayed as spheres. ECL2 and TM helices are labelled where applicable. D) RMSD of bound adenosine (ADN) (calculated with respect to initial docking pose). E) Conformational fluctuation (RMSF) of adenosine. F) Distance between TM3-TM7 (from Cα atoms of R1023.50 and Y2887.53, respectively) during MD simulations starting from the inactive crystal structure (PDB entry: 4EIY). G) Distance between TM3-TM6 (from Cα atoms of R1023.50 and E2286.30, respectively). MD simulations are performed in quadruplicate. Corresponding flat-lines show the observed distance in the active (PDB entry: 6GDG) and inactive (PDB entry: 4EIY) A2aR crystal structures.