Figure 1.
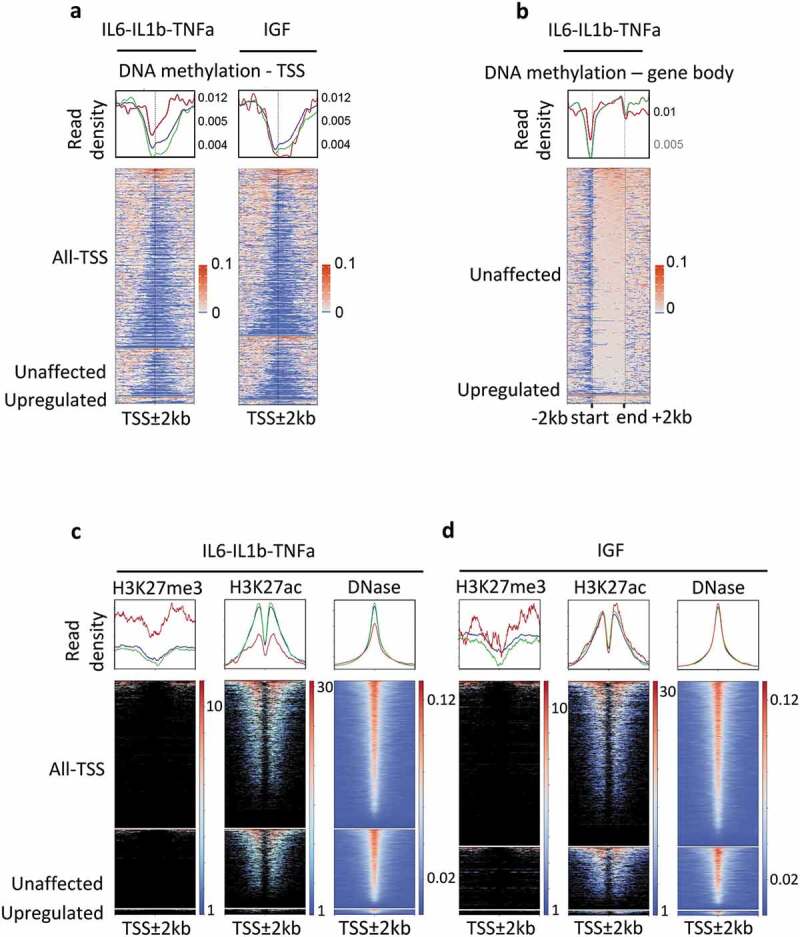
Epigenetic modifications of genes responding to the pro-inflammatory cytokines IL6, IL1b, and TNFa, in MCF7 cells.
Genes were classified as upregulated (FC ≥2, FDR < 0.05), unaffected (0.9 < FC < 1.1) and all-TSS for all expressed genes (rpkm > 0.5) when compared with the untreated MCF7 cells. IL6-IL1b-TNFa, pool of genes responding either to IL6, or IL1b, or TNFa; IGF, genes responding to IGF treatment. In red, upregulated genes; green unaffected genes; blue, all genes. a. DNA methylation density at TSS ± 2 kb regions of genes expressed, upregulated or unaffected by cytokine exposure, in MCF7; when a gene is associated with several TSS, all the TSS were kept for the construction of the heat map. b. DNA methylation density over the entire gene body of upregulated and unaffected by IL6, IL1B, and TNFa treatments, in MCF7 cells. c. Density of histones marks and DNA hypersensitivity sites within the ± 2 kb regions surrounding the TSS of genes in each subclass; IL6-IL1b-TNFa, pool of genes responding either to IL6, or IL1b, or TNFa; IGF, genes responding to IGF treatment.
