Figure 3. TNIP1 SLE risk alleles differentially affect nuclear complex binding in immune cells.
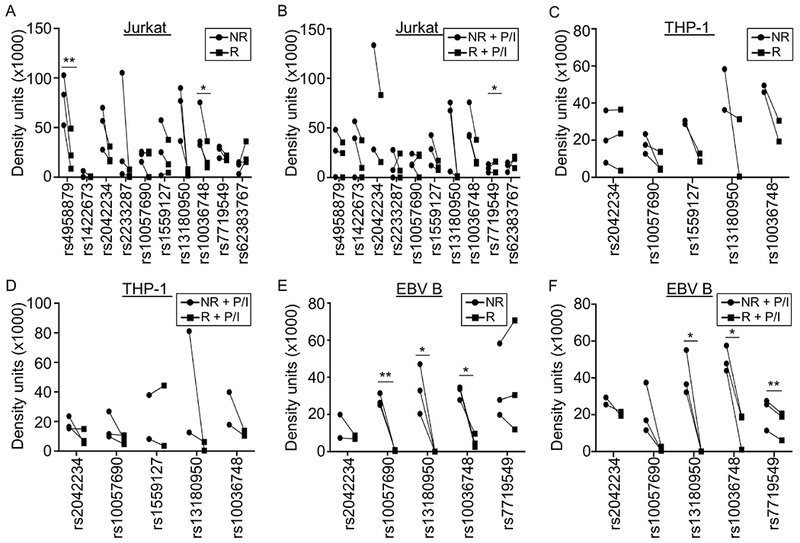
EMSAs were performed using biotinylated oligonucleotides containing the non-risk or risk alleles of the indicated variants. Nuclear extracts were from Jurkat (A,B), THP-1 (C,D), or EBV B (E,F) cells at rest (A,C,E) or after P/I stimulation (B,D,F). Statistical comparisons were performed using paired t-test; * indicates p<0.05; ** indicates p<0.01. Circles indicate non-risk allele; Squares indicate risk allele. Representative images are in Supplementary Figures 2–4.
