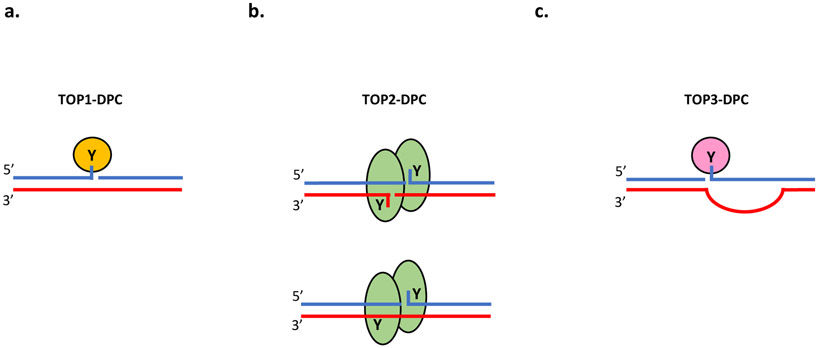
Figure 1. Architecture of TOP-DPC.
Topoisomerases cleave the DNA phosphodiester bond by carrying out nucleophilic attack of the hydroxyl end using their active tyrosine residues. This leads to formation of transient covalent linkages between the protein and the DNA, termed topoisomerase cleavage complexes (TOPcc). TOP1 enzymes attack the 3′-hydroxyl end of the DNA and cut one strand of the DNA duplex. TOP1cc can trapped by various endogenous and exogenous DNA damage and particularly by TOP1 inhibitors camptothecins. The trapped TOP1cc are referred to as TOP1 DNA-protein crosslinks (TOP1-DPC). b. Homodimeric TOP2 enzymes conduct nucleophilic attack of the 5′-hydroxyl end and cleave both strands of one DNA double helix with a 4-base stagger using Mg2+ and ATP The resulting transient TOP2cc can also be trapped by a wide range of DNA lesions and by TOP2 inhibitors such as etoposide and anthracyclines, leading to formation of TOP2-DPC. The cleavage by each subunit is not always concerted and single-strand breaks are readily produced by TOP2 enzymes, especially by their poisoning by etoposide (bottom). c. TOP3 enzymes nick one stand of single-stranded DNA by covalently linking its active tyrosine residue to the 5′ end of the DNA backbone using Mg2+ as a metal cofactor. Whether and how TOP3cc can be converted into TOP3-DPC require further investigation.