Abstract
Extended-spectrum β-lactam antimicrobials have been broadly used in food animals and humans to control infectious diseases. However, the emergence and rapid spread of extended-spectrum β-lactamase (ESBL)–producing Enterobacteriaceae, mainly Escherichia coli, have seriously threatened global health in recent decades. In this study, we determined the prevalence, antimicrobial susceptibility, and genetic properties of ESBL-producing E. coli (ESBL-EC) strains isolated from food animals in South Korea. A total of 150 fecal samples from healthy chickens (n = 34), pigs (n = 59), and cattle (n = 57) were screened from January to July 2018. Among these, 77 non-duplicate cefotaxime-resistant ESBL-EC strains were isolated from 32 chicken, 41 pig, and 4 cattle samples, with the corresponding occurrence rates of 94.1, 69.5, and 7.0%, respectively. All the isolates showed multidrug resistance (MDR) and produced at least one type of β-lactamase, including CTX-M (98.7%) and TEM (40.3%). CTX-M-14 (53.1%), CTX-M-55 (53.7%), and CTX-M-65 (50.0%) were the predominant genotypes in the chicken, pig, and cattle samples, respectively. Multilocus sequence typing revealed 46 different sequence types (STs), including the human-associated extraintestinal pathogenic E. coli ST131 (n = 2), ST10 (n = 5), ST38 (n = 1), ST410 (n = 4), ST354 (n = 2), ST58 (n = 3), ST117 (n = 1), and ST457 (n = 1). To the best of our knowledge, this is the first report of pandemic E. coli ST131 in non-human isolates in South Korea. Our results demonstrate the high prevalence and diversity of MDR-ESBL-EC in food animals and highlight them as potential pathogenic ESBL-EC reservoirs that may pose a high risk to human health.
Keywords: Escherichia coli, extended-spectrum β-lactamase, multidrug resistance, food animal, South Korea
Introduction
Extended-spectrum β-lactam antimicrobials have been widely used to treat bacterial infections both in humans and animals. Since the first extended-spectrum β-lactamase (ESBL) was described in Germany in 1983, the global spread of ESBL-producing Escherichia coli (ESBL-EC), including the pandemic E. coli sequence type (ST) 131 clone, has led to a rapid increase in the population of ESBL-EC strains worldwide (Pitout and Laupland, 2008; Rogers et al., 2011). The most widespread ESBLs are CTX-M–type β-lactamases, which can be divided into five major groups (CTX-M groups 1, 2, 8, 9, and 25) (Bonnet, 2004; Canton et al., 2012; Bevan et al., 2017), and at least 214 CTX-M variants have been detected1 accessed January 24, 2020. Among these, CTX-M-15 in the CTX-M group 1 and CTX-M-14 in the CTX-M group 9 are prevalent in most countries (Bevan et al., 2017). Likewise, both variants have been predominantly detected in clinical ESBL-EC isolates in South Korea (Song et al., 2009; Kim et al., 2019).
As ESBL-EC strains are rising in humans, they have also been increasingly isolated from food animals in different geographical regions, including China (Rao et al., 2014), Germany (Laube et al., 2013), Netherlands (Hordijk et al., 2013), Tunisia (Maamar et al., 2016), and United States (Markland et al., 2019). Moreover, multidrug-resistant (MDR) ESBL-EC pathogens, which pose a serious threat to human health due to the limited treatment options, extensively disseminate among food animals (Ho et al., 2011; Vitas et al., 2018), which are considered to be the primary reservoirs of antimicrobial-resistant enteric bacteria, although the routes of transmission to humans are unclear. Such bacteria can presumably pass through the food chain or via close contact and can colonize the intestines of humans (Carattoli, 2008). In fact, the same genetic elements and/or STs have been observed between human and food animal isolates of ESBL-EC (Moodley and Guardabassi, 2009; Leverstein-van Hall et al., 2011; Tamang et al., 2013a; Hammerum et al., 2014; Dahms et al., 2015), suggesting the possibility of clonal and genetic transmissions between these settings. Previous studies conducted in South Korea have mainly focused on the prevalence and characteristics of ESBL genes of E. coli isolates from food animals (Tamang et al., 2013b; Shin et al., 2017), but their relatedness to human-associated clonal lineages has rarely been investigated.
In this study, we evaluated the prevalence, antimicrobial susceptibility, and molecular genetic features of ESBL-EC strains isolated from food animals in South Korea. Furthermore, we assessed the epidemiological relatedness of the clonal populations to human-associated E. coli STs according to a national surveillance program.
Materials and Methods
Isolation and Identification of ESBL-EC From Food Animals
A total of 150 healthy food animals, including 34 chickens, 59 pigs, and 57 cattle, were obtained from 28, 34, and 53 farms (115 in total), respectively, across the country in South Korea. Fecal samples were collected from the intestinal tracts of individual animals slaughtered at the slaughterhouses. For E. coli isolation, 0.1 g of the samples was inoculated to 9 mL of Tryptone Soya Broth (Oxoid, Basingstoke, United Kingdom) containing 0.4 μg/mL vancomycin (Wako Pure Chemical Industries, Hyogo, Japan) and incubated at 37°C for 4 h. A loopful of each enrichment was streaked on MacConkey screening plate supplemented with 2 μg/mL cefotaxime and incubated at 37°C for 24 h. Subsequently, one pink or reddish colony suspected of comprising E. coli from each fecal sample was randomly selected using a sterile platinum loop and cultured on CHROMagar ESBL (CHROMagar, Paris, France) at 37°C for 24 h. One dark pink to reddish single colony selected from each plate was grown on Tryptone Soya Agar (Oxoid) at 37°C for 4 h, and the pure isolates were used for further characterization. The species of the isolates were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany) with score values ≥2.0. Extended-spectrum β-lactamase production was confirmed via the double-disk synergy test using disks containing amoxicillin–clavulanic acid (20/10 μg), cefotaxime (30 μg), cefepime (30 μg), and ceftazidime (30 μg).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibilities testing was performed by the disk diffusion method in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI, M100-S27) (CLSI, 2017) using commercial disks (Oxoid) supplemented with 21 antimicrobial agents as follows: gentamicin (10 μg), amikacin (30 μg), ertapenem (10 μg), imipenem (10 μg), meropenem (10 μg), cefazolin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), cefoxitin (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), tigecycline (15 μg), aztreonam (30 μg), ampicillin (10 μg), piperacillin (100 μg), amoxicillin–clavulanic acid (20/10 μg), ampicillin–sulbactam (10/10 μg), chloramphenicol (30 μg), and tetracycline (30 μg). Mueller–Hinton agar plate (Difco Laboratories, Detroit, MI, United States) was swabbed with a pure ESBL-EC suspension adjusted to a 0.5 McFarland standard. The disks were placed on the agar using a disk dispenser (Oxoid), and the plate was incubated at 37°C for 24 h. The zone diameters of growth inhibition were measured using an electronic caliper. All of the results were interpreted according to the zone diameter breakpoints of the CLSI guidelines (CLSI, 2017) except that the tigecycline results were interpreted according to the European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoint version 7.1 (EUCAST, 2017; Supplementary Table S1). Escherichia coli ATCC 25922 was used as the reference strain. Multidrug resistance was determined as non-susceptibility to at least one agent in ≥3 antimicrobial classes, and extensive drug resistance (XDR) was defined as non-susceptibility to at least one agent in all but ≤2 classes (Magiorakos et al., 2012).
Molecular Characterization of ESBL Genes
Total DNAs of ESBL-EC isolates were extracted and purified using G-spin Total DNA Extraction Kit (iNtRON Biotechnology, Seongnam, South Korea) according to the manufacturer’s instructions. To identify the ESBL alleles of the isolates, the extracts were subjected to polymerase chain reaction (PCR) analyses using the specific primer pairs, including blaTEM, blaSHV, and blaCTX–M groups 1, 2, 9, and 25 (Supplementary Table S2). After the bidirectional Sanger sequencing of the amplicons by BIOFACT (Daejeon, South Korea), the resultant sequences were compared with the published β-lactamase gene sequences from the GenBank database of the NCBI using the online BLAST program2.
Conjugation and Plasmid Typing Assay
Conjugation assay was performed with 24 blaCTX–M–55-positive donors isolated from food animals and E. coli J53-AziR recipient strains. Transconjugants were selected on MacConkey agar plates containing 2 μg/mL of cefotaxime and 100 μg/mL of sodium azide. The presence of the blaCTX–M–55 gene in the transconjugants was confirmed by PCR analysis using the CTX-M-1F/R primer pair described in Supplementary Table S2. Replicon typing of the transconjugant plasmids were tested for the major plasmid incompatibility groups among Enterobacteriaceae (HI1, HI2, I1-Iγ, X, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIAS, F, K, and B/O) using a PCR-based replicon typing method (Carattoli et al., 2005). The IncF plasmids containing FIB were further analyzed for distinguishing variants by a replicon sequence typing (RST) scheme (Supplementary Table S2) as previously described (Villa et al., 2010). The results of sequences were compared with the plasmid multilocus sequence typing (MLST) database deposited at http://pubmlst.org/plasmid/. The number of plasmids in the transconjugants was solved by pulsed field gel electrophoresis (PFGE) with S1 nuclease (Thermo Fisher Scientific, Waltham, MA, United States) digestion of total DNA using the CHEF MAPPER system (Bio-Rad Laboratories, Hercules, CA, United States).
Phylogenetic Characterization
The DNA extracts of the isolates were used for multiplex PCR targeting chuA, yjaA, and the DNA fragment TspE4.C2, as described previously (Clermont et al., 2000). Phylogroups A and B1 are typically commensal, whereas groups B2 and D are extraintestinal virulence-associated strains (Johnson et al., 2001). The ST genotypes of the isolates were determined by PCR-based sequencing of the housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) followed by MLST in accordance with the Enterobase protocol and database3 (Wirth et al., 2006). The sequences of seven housekeeping genes were aligned using Clustal W, and the phylogenetic tree was constructed using MEGA version X based on the maximum-likelihood method using Kimura’s two-parameter model with gamma distribution and invariant sites. The E. coli phylogeny was estimated by a bootstrap analysis with 1,000 replicates (Kumar et al., 2018).
Results
Prevalence and Antimicrobial Susceptibility of ESBL-EC Strains in Food Animals
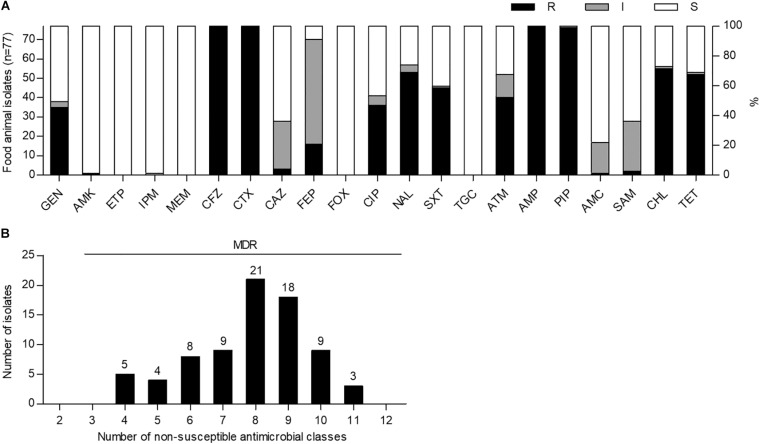
To investigate the prevalence of ESBL-EC strains in food animals, we cultured a total of 150 fecal samples from 34 chickens, 59 pigs, and 57 cattle on MacConkey agar plates containing cefotaxime (2 μg/mL) and then purified single colonies on CHROMagar ESBL plates. Among them, 77 non-duplicate cefotaxime-resistant E. coli strains were isolated from 32 chickens (94.1%), 41 pigs (69.5%), and 4 cattle (7.0%). Antimicrobial susceptibility testing of 21 antimicrobial substances from 14 classes showed that all of the isolates were not susceptible to cefotaxime (extended-spectrum cephalosporin class), cefazolin (non-extended-spectrum cephalosporin class), and ampicillin and piperacillin (penicillin class), indicating MDR (≥4 classes) phenotypes; however, they were susceptible to ertapenem, meropenem, cefoxitin, and tigecycline (Figure 1 and Supplementary Table S3). High frequencies of amikacin (76/77, 98.7%) and imipenem (76/77, 98.7%) susceptibilities were also observed. Although there was no XDR strain, three pig isolates (EC21, EC37, and EC43) showed intermediate resistance or resistance to at least one agent in all but three antimicrobial classes investigated, as described in Supplementary Tables S4, S5.
FIGURE 1.
Antimicrobial susceptibility and multidrug resistance profiles of cefotaxime-resistant Escherichia coli isolates from food animals in South Korea. (A) Antimicrobial susceptibilities were analyzed by the disk agar diffusion method. GEN, gentamicin; AMK, amikacin; ETP, ertapenem; IPM, imipenem; MEM, meropenem; CFZ, cefazolin; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; FOX, cefoxitin; CIP, ciprofloxacin; NAL, nalidixic acid; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; ATM, aztreonam; AMP, ampicillin; PIP, piperacillin; AMC, amoxicillin–clavulanic acid; SAM, ampicillin–sulbactam; CHL, chloramphenicol; TET, tetracycline; R, resistant; I, intermediate resistant; S, susceptible. (B) Multidrug resistance was determined as non-susceptibility to at least one agent in three or more antimicrobial classes. MDR, multidrug resistance.
Characterization of ESBL Genes
Next, the genotypes responsible for ESBL production were investigated using PCR-based sequencing for blaTEM, blaSHV, and blaCTX–M groups 1, 2, 9, and 25. The most prevalent ESBL genotype was represented by blaCTX–M group 1 (46/77, 59.7%), followed by blaCTX–M group 9 (31/77, 40.3%) (Table 1). The sequence identities with the β-lactamase gene sequences of the NCBI database using BLAST search were as follows: blaTEM–1 (>99.5%), blaCTX–M–1 (>99.8%), blaCTX–M–3 (99.9%), blaCTX–M–15 (>99.4%), blaCTX–M–55 (>99.3%), blaCTX–M–14 (>99.7%), and blaCTX–M–65 (>99.6%). All sequenced blaTEM amplicons belonged to non-ESBL blaTEM–1 (31/77, 40.3%), whereas blaSHV and blaCTX–M groups 2 and 25 were not detected. The predominant CTX-M–type ESBLs were CTX-M-14 in chickens (17/32, 53.1%), CTX-M-55 in pigs (22/41, 53.7%), and CTX-M-65 in cattle (2/4, 50.0%). CTX-M-1, CTX-M-3, and CTX-M-15 were also identified at various frequencies in each animal species. The ESBL gene profile of an individual isolate is given in Supplementary Table S4. In addition, we carried out the conjugation assay with azide-resistant E. coli J53 recipient strain, focusing on blaCTX–M–55. The transmission of blaCTX–M–55 gene was observed in 14 of 24 blaCTX–M–55-positive ESBL-EC isolates (Supplementary Table S6). The J53 transconjugants were further analyzed using PCR-based replicon typing. F, FIB, I1-Iγ, K, N, and FIA replicons were identified in 12, 10, 5, 3, 2, and 1 transconjugants (Supplementary Table S6). The RST discriminated F1 (9/10, 90.0%) and F20 (1/10, 10.0%) plasmids among the IncF plasmids containing FIB. S1-PFGE analysis revealed that 10 transconjugants (EC4, EC7, EC10, EC16, EC18, EC19, EC31, EC33, EC39, and EC62) contained one plasmid, whereas four (EC17, EC21, EC35, and EC40) contained two plasmids.
TABLE 1.
Extended-spectrum β-lactamase genotypes of cefotaxime-resistant Escherichia coli isolates from food animals.
| bla genotype |
Number (%) of cefotaxime-resistant E. coli isolates |
|||
| Chicken | Pig | Cattle | Total | |
| (n = 32) | (n = 41) | (n = 4) | (n = 77) | |
| TEM-1 | 10 (31.3) | 21 (51.2) | 0 | 31 (40.3) |
| CTX-M groups | 32 (100) | 40 (97.6) | 4 (100) | 76 (98.7) |
| CTX-M group 1 | 10 (31.3) | 33 (80.5) | 2 (50.0) | 45 (58.4) |
| CTX-M group 9 | 21 (65.6) | 7 (17.1) | 2 (50.0) | 30 (39.0) |
| CTX-M group 1 and group 9 | 1 (3.1) | 0 | 0 | 1 (1.3) |
| CTX-M genotypes | 5 (15.6) | |||
| CTX-M-1 | 0 | 0 | 5 (6.5) | |
| CTX-M-3 | 0 | 1 (2.4) | 0 | 1 (1.3) |
| CTX-M-15 | 4 (12.5) | 10 (24.4) | 1 (25.0) | 15 (19.5) |
| CTX-M-55 | 1 (3.1) | 22 (53.7) | 1 (25.0) | 24 (31.2) |
| CTX-M-14 | 16 (50.0) | 4 (9.8) | 0 | 20 (26.0) |
| CTX-M-65 | 5 (15.6) | 3 (7.3) | 2 (50.0) | 10 (13.0) |
| CTX-M-15 and CTX-M-14 | 1 (3.1) | 0 | 0 | 1 (1.3) |
Phylogenetic Analysis of ESBL-EC Isolates
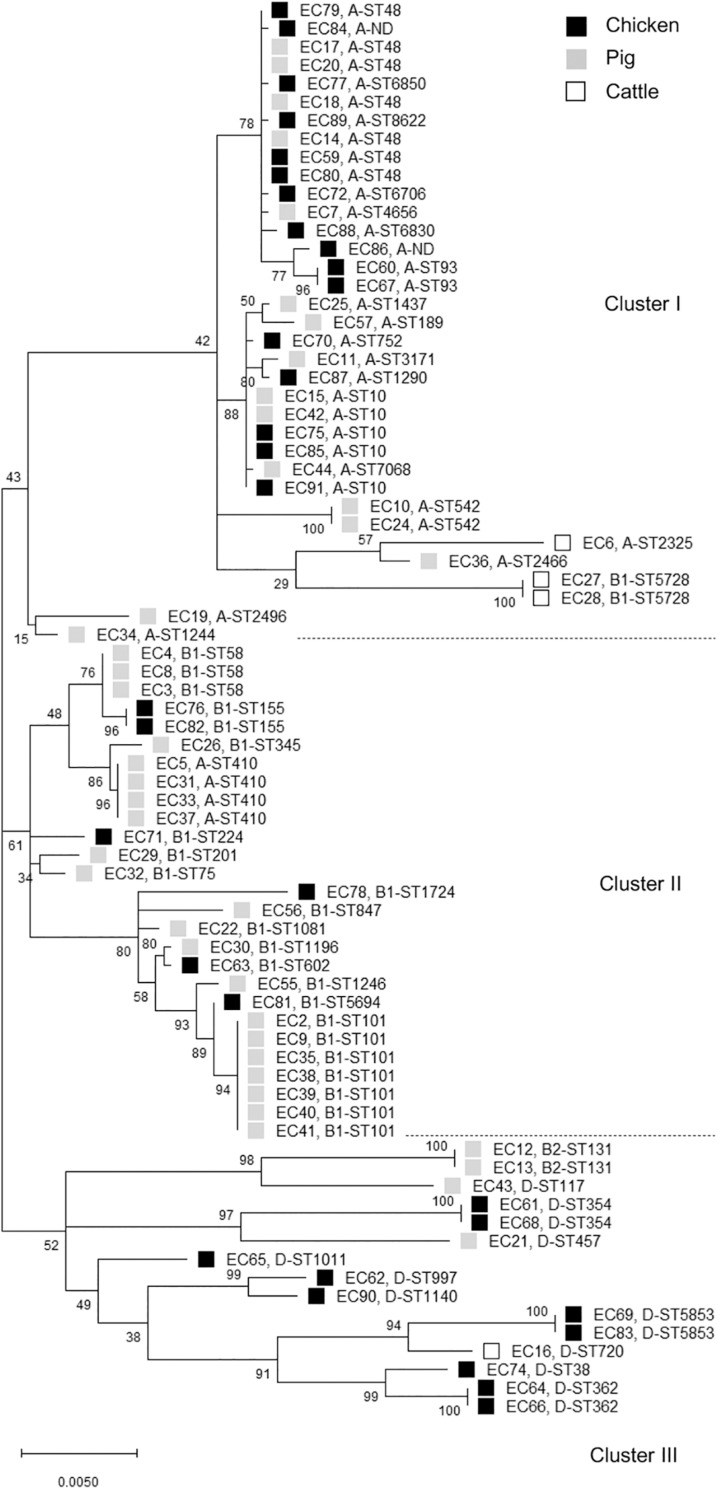
The distribution of phylogroups showed that subgroup A was predominant (37/77, 48.1%), followed by subgroups B1 (25/77, 32.5%), D (13/77, 16.9%), and B2 (2/77, 2.6%) in ESBL-EC isolates from food animals (Table 2), thus suggesting the higher distribution of commensal groups A and B1 than pathogenic groups B2 and D. However, the subgroups were differentially distributed among the animal species. The chicken isolates mainly belonged to subgroups A and D, whereas the pig isolates mostly belonged to subgroups A and B1. Through MLST analysis of a total of 77 isolates, we determined 46 distinct E. coli STs (21 chickens, 24 pigs, and 3 cattle), among which ST10 and ST48 were found in both chicken and pig isolates, and 2 unknown STs (Table 3). The proportions of STs detected only once among the isolates were 43.8% (14/32) in chickens, 41.5% (17/41) in pigs, and 50.0% (2/4) in cattle. The highest number of chicken isolates belonged to ST10 or ST48 (3/32, 9.4% each). Among the pig isolates, ST101 was the most prevalent lineage (7/41, 17.1%) and pandemic ST131 was also identified (2/41, 4.9%). The MLST-based phylogenetic tree showed that ESBL-EC isolates were branched into three major clusters, among which Cluster I and Cluster II consisted of 89.2% of subgroup A and 92% of subgroup B1, respectively, and Cluster III included all of the subgroups B2 and D (Figure 2).
TABLE 2.
Phylogroups of cefotaxime-resistant Escherichia coli isolates from food animals.
| Phylogroup |
Number (%) of cefotaxime-resistant E. coli isolates |
|||
| Chicken (n = 32) | Pig (n = 41) | Cattle (n = 4) | Total (n = 77) | |
| A | 16 (50.0) | 20 (48.8) | 1 (25.0) | 37 (48.1) |
| B1 | 6 (18.8) | 17 (41.5) | 2 (50.0) | 25 (32.5) |
| B2 | 0 | 2 (4.9) | 0 | 2 (2.6) |
| D | 10 (31.3) | 2 (4.9) | 1 (25.0) | 13 (16.9) |
TABLE 3.
MLST analysis of cefotaxime-resistant Escherichia coli isolates from food animals.
| MLST |
Number (%) of cefotaxime-resistant E. coli isolates |
|||
| Chicken | Pig | Cattle | Total | |
| (n = 32) | (n = 41) | (n = 4) | (n = 77) | |
| ST10 | 3 (9.4) | 2 (4.9) | 0 | 5 (6.5) |
| ST48 | 3 (9.4) | 4 (9.8) | 0 | 7 (9.1) |
| ST58 | 0 | 3 (7.3) | 0 | 3 (3.9) |
| ST93 | 2 (6.3) | 0 | 0 | 2 (2.6) |
| ST101 | 0 | 7 (17.1) | 0 | 7 (9.1) |
| ST131 | 0 | 2 (4.9) | 0 | 2 (2.6) |
| ST155 | 2 (6.3) | 0 | 0 | 2 (2.6) |
| ST354 | 2 (6.3) | 0 | 0 | 2 (2.6) |
| ST362 | 2 (6.3) | 0 | 0 | 2 (2.6) |
| ST410 | 0 | 4 (9.8) | 0 | 4 (5.2) |
| ST542 | 0 | 2 (4.9) | 0 | 2 (2.6) |
| ST5728 | 0 | 0 | 2 (50.0) | 2 (2.6) |
| ST5853 | 2 (6.3) | 0 | 0 | 2 (2.6) |
| Once detected ST | 14 (43.8) | 17 (41.5) | 2 (50.0) | 33 (42.9) |
| Not determined ST | 2 (6.3) | 0 | 0 | 2 (2.6) |
ST, sequence type.
FIGURE 2.
Phylogenetic tree of cefotaxime-resistant Escherichia coli isolates from chickens, pigs, and cattle. The maximum likelihood phylogenetic tree was constructed using Mega X software based on the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) and Kimura’s 2-parameter model. Bootstrap support percentages (1,000 replicates) were indicated in the different branches. Scale bar at the bottom represents the genetic distance. The phylogroup and sequence type (ST) of each isolate were displayed. Black, gray, and white squares represent chicken, pig, and cattle, respectively. ND, not determined ST.
Discussion
The prevalence of ESBL-EC in food animals (94.1% in chickens, 69.5% in pigs, and 7.0% in cattle) in this study was found to be higher than those reported in previous investigations before and after 2010 (33.3% in chickens, 21.5% in pigs, and 0.2% in cattle) (Tamang et al., 2013b; Lim et al., 2015) in South Korea. In the susceptibility testing for 14 antimicrobial classes, all of the isolates showed the MDR phenotypes with a resistance range of 4–11 classes (Figure 1B). In general, ESBLs can hydrolyze extended-spectrum cephalosporins and monobactams but not carbapenems and cephamycins and are inhibited by β-lactamase inhibitors (Canton et al., 2012). Consistent with these properties, ESBL-EC isolates in this study showed a relatively high susceptibility to ertapenem, imipenem, meropenem, cefoxitin, amoxicillin − clavulanic acid, and ampicillin − sulbactam (Figure 1A and Supplementary Table S3). In addition, ESBL-producing Enterobacteriaceae among clinical isolates have been reported to be mostly susceptible to tigecycline and amikacin (Morosini et al., 2006; Denisuik et al., 2019), and similarly, the same phenotype was also found in food animal isolates in this study (Figure 1A and Supplementary Table S3).
The epidemiology of CTX-M β-lactamases has been globally changed (Bevan et al., 2017). CTX-M-55, which differs from CTX-M-15 by one nucleotide at 239 resulting in A77V substitution, displayed enhanced cephalosporin-hydrolyzing activity and structural stability (He et al., 2015). The population of CTX-M-55–producing ESBL-EC strains in China is showing increasing trends in both human and food animals (Hu et al., 2013; Rao et al., 2014; Zhang et al., 2014). In South Korea, CTX-M-15 and CTX-M-14 had been reported to be the predominant CTX-M β-lactamases in ESBL-EC isolates from food animals (Tamang et al., 2013b; Shin et al., 2017), however, CTX-M-55 was most prevalently detected in companion animals (Hong et al., 2019) and raw retail chickens (Park et al., 2019). In this study, we found that food animal ESBL-EC predominantly produced the CTX-M-55 enzyme (Table 1), suggesting that the CTX-M-55 may be supplanting CTX-M-15. The blaCTX–M–55 gene associated with cefotaxime-resistant phenotype was transferable from 14 of 24 blaCTX–M–55–positive ESBL-EC isolates to other E. coli strain by conjugation as described in Supplementary Table S6, suggesting that food animals may acquire blaCTX–M–55 through a A77V substitution from blaCTX–M–15 but also horizontal gene transfer. Plasmids play a critical role in the global dissemination of ESBL genes (Wang et al., 2018). The blaCTX–M–55 gene was frequently found on IncF, IncI1, and IncHI2 plasmids of E. coli in many countries, including China (Yang et al., 2014; Wang et al., 2018), France (Lupo et al., 2018), and United States (McGann et al., 2016). Similarly, the presence of I1-Iγ, F, and P plasmids in blaCTX–M–55–positive ESBL-EC isolates from pigs has been documented in South Korea (Tamang et al., 2013b). This study showed that most of the blaCTX–M–55–positive transconjugants (12/14, 85.7%) carried IncF replicon in combination with other types, including FIB, I1-Iγ, K, N, and/or FIA. The diversity of plasmid types was increased in comparison with those in previous report (Tamang et al., 2013b), which may reflect more influx of various antimicrobial-resistant genes. Among IncF plasmids in the blaCTX–M–55-positive E. coli isolates from animals in China, F33 plasmids were the most prevalent replicon STs (Yang et al., 2015). In contrast, F1 types, which are commonly found to carry ESBL genes (Brolund, 2014), were most predominantly identified in this study.
In this study, the food animal ESBL-EC isolates mainly belonged to commensal groups A or B1. Most of each phylogroup to which they belonged was allocated to a distinct cluster, whereas there was no difference between animal species allocations (Figure 2), suggesting that the phylogenetic relationships among the ESBL-EC STs may be closely related to their phylogroups regardless of the host animal species. Notably, the different breed composition of cattle has been reported to be associated with gut microbiota structure and β-lactam resistance (Fan et al., 2019), suggesting the impact of animal genetics on the antimicrobial-resistant bacteria profile even within the same species. We further detected various clonal STs in these isolates by MLST analysis. The rate of STs detected only once was 42.9%, suggesting that food animals possess a wider variety of MDR-ESBL-EC STs. Interestingly, among them, ST131, ST10, ST38, ST410, ST354, ST58, and ST117 have been reported as major extraintestinal pathogenic E. coli (ExPEC) lineages, which cause an extraintestinal infection in human (Manges et al., 2019). Rarely reported ST457 also belongs to ExPEC (Seni et al., 2018). The globally predominant ExPEC ST131 belongs to the highly virulent phylogroup B2 and causes both community-onset and hospital-onset infections (Nicolas-Chanoine et al., 2014). It commonly produces ESBLs and is highly associated with MDR, including resistance to fluoroquinolone. The epidemiology and characteristics of the ST131 clonal group have mostly been investigated in human clinical isolates, and animal and environmental clones have been identified only in a few studies worldwide. Similarly in South Korea, E. coli ST131 was found to be the most prevalent clone in patients with urinary tract infections and bacteremia and commonly harbored blaCTX–M–15 and blaCTX–M–14 (Lee et al., 2010; Cha et al., 2016; Kim H. et al., 2017, Kim Y.A. et al., 2017). There have been few reports on E. coli ST131 from food animals (Nicolas-Chanoine et al., 2014). To the best of our knowledge, this is the first report of the presence of E. coli ST131 in food animals in South Korea. We identified two pig E. coli ST131 isolates, which harbored both blaCTX–M–65 and blaTEM–1 genes and had MDR phenotypes. Human E. coli ST131 carrying both blaCTX–M–65 and blaTEM–1 has been detected in Germany (Cullik et al., 2010). Clonal populations of ST410 are present in humans, companion animals, livestock, and the environment (Falgenhauer et al., 2016) and pose a high risk of causing ExPEC outbreaks in hospitals worldwide (Roer et al., 2018). ST10, ST58, and ST117 lineages have also been detected in both humans and food animals (Manges et al., 2015; McKinnon et al., 2018). In addition, ST38 and ST101 were present in chicken and pig samples analyzed in this study, respectively, which are more related to hospital-onset than to community-onset infections in South Korea (Yoo et al., 2013; Kim et al., 2016). Extended-spectrum β-lactamase genes may circulate among food animals, farm workers, and the farm environment (Tamang et al., 2013a), but also cefotaxime-resistant bacteria can be isolated from food animals raised without cephalosporins (Mir et al., 2016, 2018), suggesting the animal acquisition of antimicrobial resistance from the environment. Together, these results suggest the two-way spread of resistant bacteria: food animals may be getting them from humans, hospital waste, and the environment or from their feed and fodder.
In conclusion, our results demonstrate the increasing occurrence and clonal diversity of MDR-ESBL-EC strains in food animals. These strains include pathogenic human-associated lineages, such as the E. coli ST131 clone. To explore the possible origin of the two ST131 strains found in pigs, it would be interesting to compare their core genome sequences with those of other ST131 from humans in South Korea. Given the possibility of direct transmission of antimicrobial resistance to humans through the food chain, this study also demonstrates the importance of understanding the dynamics of MDR E. coli in food animals.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the animal study because we used livestock feces from slaughterhouses, not livestock itself.
Author Contributions
S-SO and JSh contributed conception and design of the study. JSo, S-SO, and JK collected the samples and performed the experiments. JSh wrote the manuscript. All authors analyzed the data, contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Drs. Junyoung Kim, Young Ah Kim, and Hyukmin Lee for their helpful discussions and the Research Institute of Bacterial Resistance at Yonsei University College of Medicine for providing services.
Funding. This research was supported by the Korea Centers for Disease Control and Prevention (2017ER540301), the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2018R1A6A1A03025523), and Inha University Research Grant (2018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00604/full#supplementary-material
References
- Bevan E. R., Jones A. M., Hawkey P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72 2145–2155. 10.1093/jac/dkx146 [DOI] [PubMed] [Google Scholar]
- Bonnet R. (2004). Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48 1–14. 10.1128/aac.48.1.1-14.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brolund A. (2014). Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect. Ecol. Epidemiol. 4:24555 10.3402/iee.v4.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R., Gonzalez-Alba J. M., Galan J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110 10.3389/fmicb.2012.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2008). Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14 117–123. 10.1111/j.1469-0691.2007.01851.x [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Cha M. K., Kang C. I., Kim S. H., Cho S. Y., Ha Y. E., Wi Y. M., et al. (2016). Comparison of the microbiological characteristics and virulence factors of ST131 and non-ST131 clones among extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia. Diagn. Microbiol. Infect. Dis. 84 102–104. 10.1016/j.diagmicrobio.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66 4555–4558. 10.1128/aem.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2017). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cullik A., Pfeifer Y., Prager R., von Baum H., Witte W. (2010). A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 59 580–587. 10.1099/jmm.0.016188-0 [DOI] [PubMed] [Google Scholar]
- Dahms C., Hubner N. O., Kossow A., Mellmann A., Dittmann K., Kramer A. (2015). Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania Germany. PLoS One 10 e0143326 10.1371/journal.pone.0143326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisuik A. J., Karlowsky J. A., Adam H. J., Baxter M. R., Lagace-Wiens P. R. S., Mulvey M. R., et al. (2019). Dramatic rise in the proportion of ESBL-producing Escherichia coli and Klebsiella pneumoniae among clinical isolates identified in Canadian hospital laboratories from 2007 to 2016. J. Antimicrob. Chemother. 74 64–71. 10.1093/jac/dkz289 [DOI] [PubMed] [Google Scholar]
- EUCAST (2017). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. Sweden: EUCAST. [Google Scholar]
- Falgenhauer L., Imirzalioglu C., Ghosh H., Gwozdzinski K., Schmiedel J., Gentil K., et al. (2016). Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 47 457–465. 10.1016/j.ijantimicag.2016.03.019 [DOI] [PubMed] [Google Scholar]
- Fan P., Nelson C. D., Driver J. D., Elzo M. A., Jeong K. C. (2019). Animal breed composition is associated with the hindgut microbiota structure and β-lactam resistance in the multibreed Angus-Brahman herd. Front. Microbiol. 10:1846 10.3389/fmicb.2019.01846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum A. M., Larsen J., Andersen V. D., Lester C. H., Skovgaard Skytte T. S., Hansen F., et al. (2014). Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 69 2650–2657. 10.1093/jac/dku180 [DOI] [PubMed] [Google Scholar]
- He D., Chiou J., Zeng Z., Liu L., Chen X., Zeng L., et al. (2015). Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 59 5976–5983. 10.1128/AAC.04920-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. L., Chow K. H., Lai E. L., Lo W. U., Yeung M. K., Chan J., et al. (2011). Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ‘critically important’ antibiotics among food animals in Hong Kong, 2008-10. J. Antimicrob. Chemother. 66 765–768. 10.1093/jac/dkq539 [DOI] [PubMed] [Google Scholar]
- Hong J. S., Song W., Park H. M., Oh J. Y., Chae J. C., Shin S., et al. (2019). Clonal spread of extended-spectrum cephalosporin-resistant Enterobacteriaceae between companion animals and humans in South Korea. Front. Microbiol. 10:1371 10.3389/fmicb.2019.01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordijk J., Wagenaar J. A., van de Giessen A., Dierikx C., van Essen-Zandbergen A., Veldman K., et al. (2013). Increasing prevalence and diversity of ESBL/AmpC-type β-lactamase genes in Escherichia coli isolated from veal calves from 1997 to 2010. J. Antimicrob. Chemother. 68 1970–1973. 10.1093/jac/dkt132 [DOI] [PubMed] [Google Scholar]
- Hu Y. Y., Cai J. C., Zhou H. W., Chi D., Zhang X. F., Chen W. L., et al. (2013). Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 79 5988–5996. 10.1128/AEM.01740-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Delavari P., Kuskowski M., Stell A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183 78–88. 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- Kim H., Kim Y. A., Park Y. S., Choi M. H., Lee G. I., Lee K. (2017). Risk Factors and Molecular features of sequence type (ST) 131 extended-spectrum β-lactamase-producing Escherichia coli in community-onset bacteremia. Sci. Rep. 7:14640 10.1038/s41598-017-14621-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. A., Kim J. J., Kim H., Lee K. (2017). Community-onset extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 at two Korean community hospitals: the spread of multidrug-resistant E. coli to the community via healthcare facilities. Int. J. Infect. Dis. 54 39–42. 10.1016/j.ijid.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Kim K. G., Jeong J., Kim M. J., Park D. W., Shin J. H., Park H. J., et al. (2019). Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type β-lactamases and carbapenemases among diarrhoeagenic Escherichia coli isolates from children in Gwangju Korea: 2007-16. J. Antimicrob. Chemother. 74 2181–2187. 10.1093/jac/dkz175 [DOI] [PubMed] [Google Scholar]
- Kim S., Sung J. Y., Cho H. H., Kwon K. C., Koo S. H. (2016). Characteristics of the molecular epidemiology of CTX-M-Producing Escherichia coli isolated from a tertiary hospital in Daejeon Korea. J. Microbiol. Biotechnol. 26 1643–1649. 10.4014/jmb.1603.03063 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube H., Friese A., von Salviati C., Guerra B., Kasbohrer A., Kreienbrock L., et al. (2013). Longitudinal monitoring of extended-spectrum-β-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 79 4815–4820. 10.1128/AEM.00856-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Y., Choi H. J., Choi J. Y., Song M., Song Y., Kim S. W., et al. (2010). Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J. Infect. 60 146–153. 10.1016/j.jinf.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A., Dierikx C. M., Cohen Stuart J., Voets G. M., van den Munckhof M. P., van Essen-Zandbergen A., et al. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- Lim J. S., Choi D. S., Kim Y. J., Chon J. W., Kim H. S., Park H. J., et al. (2015). Characterization of Escherichia coli-producing extended-spectrum β-lactamase (ESBL) isolated from chicken slaughterhouses in South Korea. Foodborne Pathog. Dis. 12 741–748. 10.1089/fpd.2014.1921 [DOI] [PubMed] [Google Scholar]
- Lupo A., Saras E., Madec J. Y., Haenni M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73 867–872. 10.1093/jac/dkx489 [DOI] [PubMed] [Google Scholar]
- Maamar E., Hammami S., Alonso C. A., Dakhli N., Abbassi M. S., Ferjani S., et al. (2016). High prevalence of extended-spectrum and plasmidic AmpC β-lactamase-producing Escherichia coli from poultry in Tunisia. Int. J. Food Microbiol. 231 69–75. 10.1016/j.ijfoodmicro.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Manges A. R., Geum H. M., Guo A., Edens T. J., Fibke C. D., Pitout J. D. D. (2019). Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32:CMR.00135-18 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A. R., Harel J., Masson L., Edens T. J., Portt A., Reid-Smith R. J., et al. (2015). Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog. Dis. 12 302–310. 10.1089/fpd.2014.1860 [DOI] [PubMed] [Google Scholar]
- Markland S., Weppelmann T. A., Ma Z., Lee S., Mir R. A., Teng L., et al. (2019). High prevalence of cefotaxime resistant bacteria in grazing beef cattle: a cross sectional study. Front. Microbiol. 10:176 10.3389/fmicb.2019.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann P., Snesrud E., Maybank R., Corey B., Ong A. C., Clifford R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60 4420–4421. 10.1128/AAC.01103-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon J., Roy Chowdhury P., Djordjevic S. P. (2018). Genomic analysis of multidrug-resistant Escherichia coli ST58 causing urosepsis. Int. J. Antimicrob. Agents 52 430–435. 10.1016/j.ijantimicag.2018.06.017 [DOI] [PubMed] [Google Scholar]
- Mir R. A., Weppelmann T. A., Johnson J. A., Archer D., Morris J. G., Jr., et al. (2016). identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS One 11:e0163279 10.1371/journal.pone.0163279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir R. A., Weppelmann T. A., Teng L., Kirpich A., Elzo M. A., Driver J. D., et al. (2018). Colonization dynamics of cefotaxime resistant bacteria in beef cattle raised without cephalosporin antibiotics. Front. Microbiol. 9:500 10.3389/fmicb.2018.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley A., Guardabassi L. (2009). Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 53 1709–1711. 10.1128/AAC.01014-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosini M. I., Garcia-Castillo M., Coque T. M., Valverde A., Novais A., Loza E., et al. (2006). Antibiotic coresistance in extended-spectrum-β-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob. Agents Chemother. 50 2695–2699. 10.1128/AAC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas-Chanoine M. H., Bertrand X., Madec J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27 543–574. 10.1128/CMR.00125-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Kim J., Ryu S., Jeon B. (2019). Predominance of blaCTX-M-65 and blaCTX-M-55 in extended-spectrum β-lactamase-producing Escherichia coli from raw retail chicken in South Korea. J. Glob. Antimicrob. Resist. 17 216–220. 10.1016/j.jgar.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet. Microbiol. 172 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Roer L., Overballe-Petersen S., Hansen F., Schonning K., Wang M., Roder B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18 10.1128/mSphere.00337-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. A., Sidjabat H. E., Paterson D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 66 1–14. 10.1093/jac/dkq415 [DOI] [PubMed] [Google Scholar]
- Seni J., Peirano G., Okon K. O., Jibrin Y. B., Mohammed A., Mshana S. E., et al. (2018). The population structure of clinical extra-intestinal Escherichia coli in a teaching hospital from Nigeria. Diagn. Microbiol. Infect. Dis. 92 46–49. 10.1016/j.diagmicrobio.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Shin S. W., Jung M., Won H. G., Belaynehe K. M., Yoon I. J., Yoo H. S. (2017). Characteristics of transmissible CTX-M- and CMY-Type β-lactamase-producing Escherichia coli isolates collected from pig and chicken farms in South Korea. J. Microbiol. Biotechnol. 27 1716–1723. 10.4014/jmb.1610.10006 [DOI] [PubMed] [Google Scholar]
- Song W., Lee H., Lee K., Jeong S. H., Bae I. K., Kim J. S., et al. (2009). CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum β-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 58 261–266. 10.1099/jmm.0.004507-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M. D., Nam H. M., Gurung M., Jang G. C., Kim S. R., Jung S. C., et al. (2013a). Molecular characterization of CTX-M β-lactamase and associated addiction systems in Escherichia coli circulating among cattle, farm workers, and the farm environment. Appl. Environ. Microbiol. 79 3898–3905. 10.1128/AEM.00522-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M. D., Nam H. M., Kim S. R., Chae M. H., Jang G. C., Jung S. C., et al. (2013b). Prevalence and molecular characterization of CTX-M β-lactamase-producing Escherichia coli isolated from healthy swine and cattle. Foodborne Pathog. Dis. 10 13–20. 10.1089/fpd.2012.1245 [DOI] [PubMed] [Google Scholar]
- Villa L., Garcia-Fernandez A., Fortini D., Carattoli A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65 2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- Vitas A. I., Naik D., Perez-Etayo L., Gonzalez D. (2018). Increased exposure to extended-spectrum β-lactamase-producing multidrug-resistant Enterobacteriaceae through the consumption of chicken and sushi products. Int. J. Food Microbiol. 269 80–86. 10.1016/j.ijfoodmicro.2018.01.026 [DOI] [PubMed] [Google Scholar]
- Wang J., Zeng Z. L., Huang X. Y., Ma Z. B., Guo Z. W., Lv L. C., et al. (2018). Evolution and comparative genomics of F33:A-:B- plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere 3:e00137-18 10.1128/mSphere.00137-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. E., Sun J., Li L., Deng H., Liu B. T., Fang L. X., et al. (2015). IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front. Microbiol. 6:964 10.3389/fmicb.2015.00964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liu W., Liu Y., Wang J., Lv L., Chen X., et al. (2014). F33:A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front. Microbiol. 5:688 10.3389/fmicb.2014.00688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J. S., Kim H. M., Koo H. S., Yang J. W., Yoo J. I., Kim H. S., et al. (2013). Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J. Antimicrob. Chemother. 68 2170–2172. 10.1093/jac/dkt126 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zheng B., Zhao L., Wei Z., Ji J., Li L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659 10.1186/s12879-014-0659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.