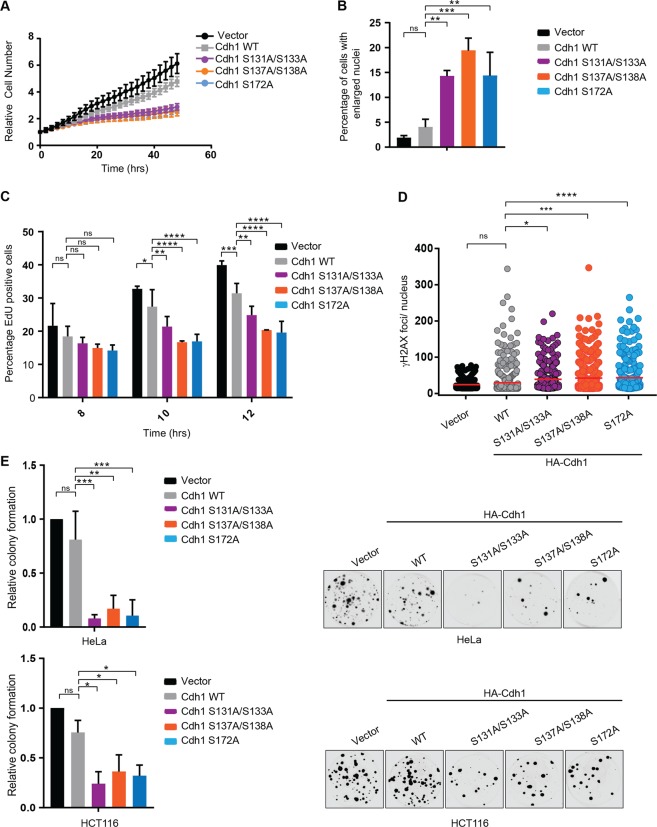
Fig. 5. Chk1-mediated phosphorylation of Cdh1 ensures proper cell-cycle progression.
a Mutation of the Chk1-mediated Cdh1 phosphorylation sites inhibits proliferation. HeLa cells were transfected with histone H2B-GFP and either vector or the indicated Cdh1 constructs. After 24 h, proliferation of the GFP-positive cells was analyzed over time. b Mutation of the Chk1-mediated Cdh1 phosphorylation sites promotes aberrant nuclear size. HeLa cells were transfected as in A. After 48 h, the percent of GFP-positive cells with enlarged nuclei was determined. Data (a, b) are represented as mean ± SD, n = 3 biological replicates **p < 0.01, ***p < 0.001 were calculated with 1-way ANOVA with Sidak’s post-test. c Mutation of the Chk1-mediated Cdh1 phosphorylation sites delays S-phase entry. Hela cells (as in a) were synchronized in mitosis with a thymidine-nocodazole block following transfection with the indicated Cdh1 constructs. After release from nocodazole, cells were pulsed with EdU for 15 min, at the indicated time points fixed and analyzed for EdU incorporation. The plot represents the percent of GFP-positive, EdU-positive nuclei. Data are represented as mean ± SD, n = 3 biological replicates and *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 were calculated with 2-way ANOVA with Dunnet’s post-test. d Mutation of the Chk1-mediated phosphorylation sites in Cdh1 induces DNA damage. The number of γH2AX foci in cells. U2OS cells as in (a) were analyzed for γH2AX foci. The graph shows the number of γH2AX foci per GFP-positive nucleus. Mean is indicated by the red line. n > 275; *p < 0.05, ***i < 0.001, ****p < 0.0001 were calculated with 1-way ANOVA with Dunnet’s post-test. e Effects of non-degradable Cdh1 on colony-forming capacity of HeLa and HCT116 cells. Cells were transfected with the indicated Cdh1 constructs. The graph represents the relative fraction of number of colonies transfected with each indicated Cdh1 constructs and normalized to vector control. n = 3 biological replicates (with three technical replicates per biological replicate), error bars represents standard deviation and *p < 0.05, **p < 0.01, ***p < 0.001 were calculated with 1-way ANOVA with Sidak’s post-test. Representative images of colony formation following each treatment are shown for both HeLa and HCT116 cells.