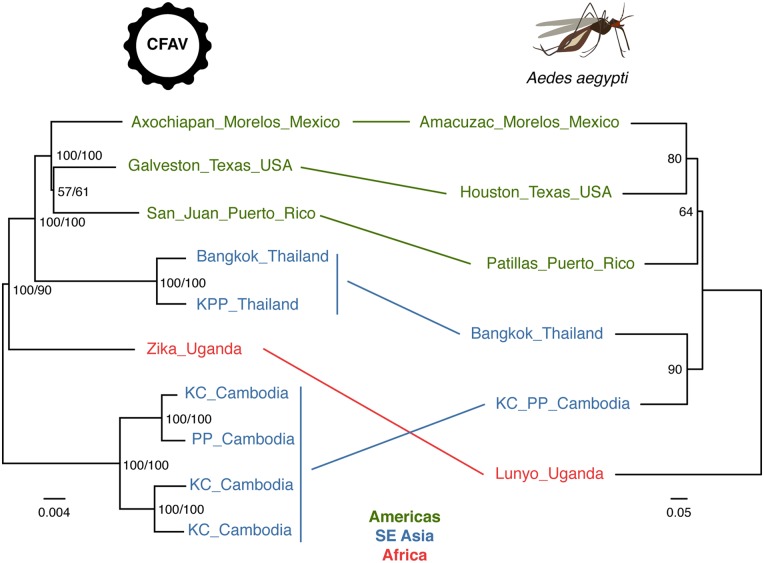
Figure 3.
Lack of phylogenetic congruence between CFAV and A.aegypti in Southeast Asia. Left: consensus tree of 1,000 ultrafast bootstrap replicate ML trees represents phylogenetic relationships between CFAV sequences based on the nearly full ORF alignment. The tree is midpoint rooted. Node support values include both ultrafast bootstrap proportion (%, first value) and posterior probability derived from the Bayesian maximum clade credibility tree (%, second value). The scale bar represents branch length in substitutions per site. Right: midpoint rooted consensus tree of 1,000 bootstrap replicates trees based on the Edwards’ genetic distances calculated from microsatellite allele frequencies among A.aegypti populations. Node support values represent bootstrap proportions (%). Lines between the trees connect the tips sharing the same geographic origin.