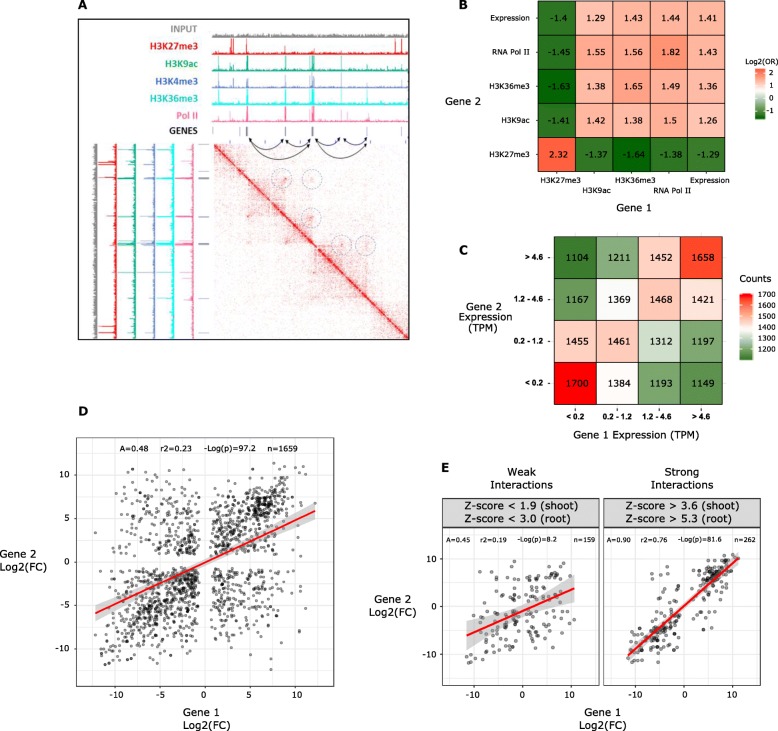
Fig. 4.
Chromatin loops define local-scale functional units of wheat genome architecture. a Integration of Hi-C and ChIP-seq data. A 2D heatmap with the interaction frequency in region chr3B-334500000-337000000 is presented. Black bars represent genes. Dashed circles represent the chromatin loop interactions. b Heatmap presenting the logarithm of odds ratios of all combinations of features of interacting genes (see the “Results” section). Positive log2(odds ratio) indicates enrichment and negative indicates depletion. c Heatmap presenting the gene expression levels of gene-to-gene loop (GGL) gene pairs. Within each gene pair, each gene was classified into a quartile of expression, and then the number of gene pairs with each combination of quartiles was counted. d Scatterplot of log2(shoot/root fold change) for pairs of interacting genes associated with a GGL conserved in shoots and roots. Loops (significant interactions) were identified from Hi-C data. e Scatterplots of log2(shoot/root fold change) for two subsets of loops: weak (first quartile of Z-score) and strong (last quartile of Z-score)