Figure 2.
Capsular switching and recombination breakpoints, pneumococcal isolates, south-western Europe
BLAST: basic local alignment search tool; ENA: European Nucleotide Archive; MLST: multilocus sequence typing; NCBI: National Center for Biotechnology Information; SNP: single nucleotide polymorphism; ST: sequence type.
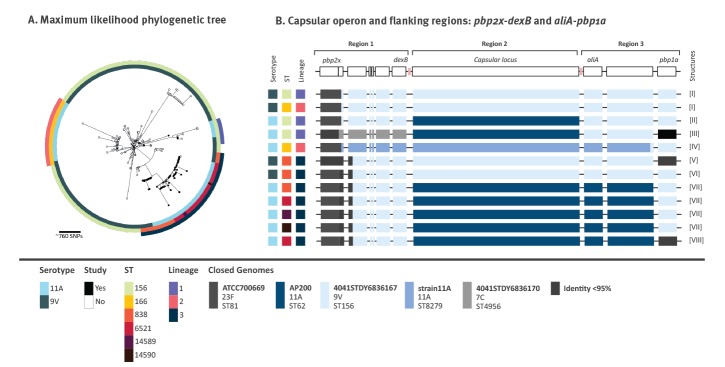
Panel A: The maximum likelihood phylogenetic tree includes the 61 sequenced strains and 238 strains of serotypes 9V and 11A for which genomes were available in ENA and which belonged to ST related to CC156 (ST156, ST166, ST838 or ST6521). The colour of each circle represents different characteristics of the strains. Outer circle: lineages; middle circle: ST; inner circle: serotypes.
Panel B: The analysis of the capsular operon was done with 61 genomes from this study and a selection of 59 from the 238 ENA genomes. The different structures are numbered I to VIII. Colours are related to the closest fully closed genome on NCBI (indicated in the legend with serotype and MLST). Black regions: areas with identity < 95% with NCBI fully closed genomes after BLAST search; pink squares: non-analysed flanking transposases.