Figure 4.
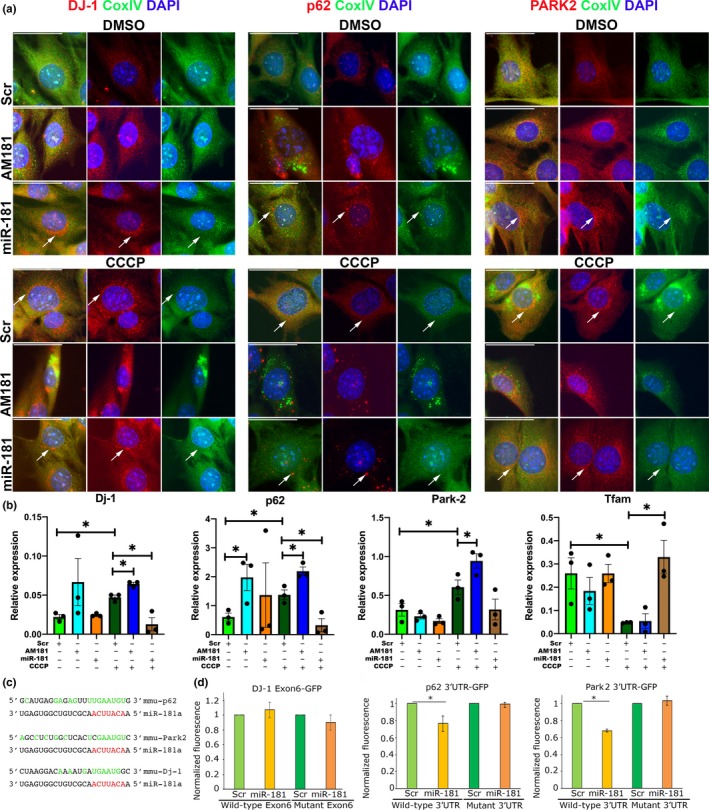
miR‐181a targets p62 and Park2 and DJ‐1 in C2C12 myoblasts. (a) Immunostaining for CoxIV (mitochondrial marker) and mitophagy‐associated genes: p62, DJ‐1, and Park2, showing accumulation of these proteins in C2C12 myoblasts following inhibition of miR‐181a function and turnover of mitochondria following overexpression of miR‐181a. Arrows indicate regions with protein expression and reduced CoxIV expression indicating mitophagy. Representative images are shown. Scale bars indicate 50 µm. (b) qPCR showing the effects of miR‐181 in C2C12 myoblasts following mitophagy induction on the expression of mitophagy‐associated genes, relative to β2‐microglobulin. n = 3–4. Error bars show SEM. *p < .05; unpaired Student's t test. (c) miR‐181a predicted binding sites within the 3′UTRs of murine p62 and Park2 and within an exon of murine DJ‐1. Red—microRNA seed sequence; green—nucleotides complementary to the microRNAs sequence. (d) Normalized GFP fluorescence in myoblasts following co‐transfection with GFP sensor construct containing wild‐type or mutated miR‐181a predicted binding site and miR‐181a mimic or scrambled miRNA mimic. n = 3. Error bars show stdev. *p < .05 Student's t test
