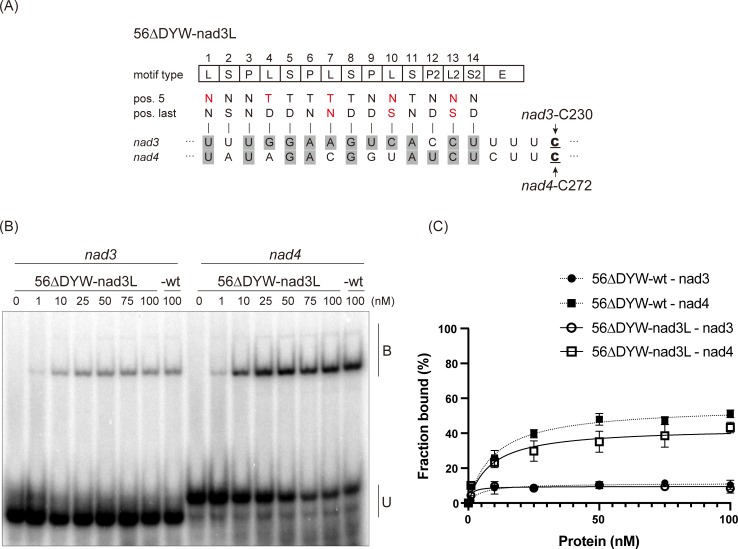
Fig 2. In vitro RNA binding assay for the PpPPR_56 variant.
(A) Alignment of the 56ΔDYW-nad3L variant to nad3 and nad4 RNAs. The key amino acids at positions 5 and last of each PPR motif are indicated. Altered amino acid residues are shown in red. The editing sites C are underlined. The aligned nucleotides are shaded in gray to indicate the matches to the proposed RNA recognition codes. (B) REMSA was performed using recombinant 56ΔDYW-nad3L, 56ΔDYW-wt, and 32P-labeled RNAs (nad3 and nad4). The recombinant protein concentrations are shown above each lane. B and U indicate the bound and unbound probe, respectively. (C) Binding curves representing the fraction of the bound probe indicated in Figs 1B and 2B. Error bars indicate ± SD (n = 3).