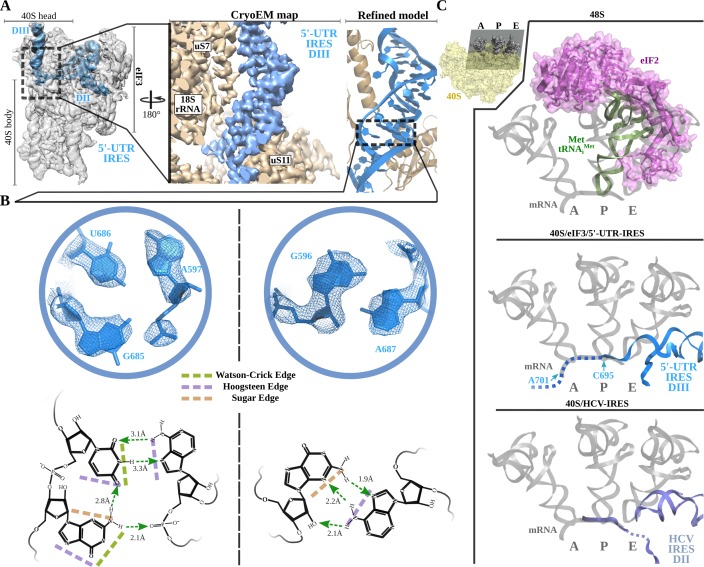
Figure 4. Non-canonical base pairing in 5'-UTR-IRES domain III assists on P-site access.
A) Overview of the 40S–5'-UTR-IRES–eIF3 cryo-EM map with 40S and eIF3 colored gray and 5'-UTR-IRES blue. On the right, a detailed view of the E-site, where 5'-UTR-IRES domain III is placed, shows the cryo-EM map with the 5'-UTR-IRES colored blue and 40S components brown. On the far right, the final refined model is colored following the same color scheme. Ribosomal proteins uS7 and uS11 as well as several 18S rRNA bases contact 5'-UTR-IRES domain III. (B) Non-canonical base pairs found in 5'-UTR-IRES domain III induce a distortion of the double helix near the E-site. At the top, two examples are shown with the refined model inserted in the experimental cryo-EM density. The corresponding chemical diagrams below show the base edges and the hydrogen bonds involved in interactions. (C) Superpositions of the canonical 48S complex (PDB ID 6FEC, top) with the HCV-IRES/40S complex (PDB ID 5A2Q, bottom) and the 40S/5'-UTR-IRES/eIF3 (middle) models, focused on the tRNA A, P and E binding sites. 5'-UTR-IRES domain III and HCV-IRES occupies a space on the E-site that overlaps with the position described for eIF2 in the canonical 48S complex. In the middle, the last residue of 5'-UTR-IRES is indicated (C695), as is the putative path along the mRNA binding channel followed by the IRES (dashed line).