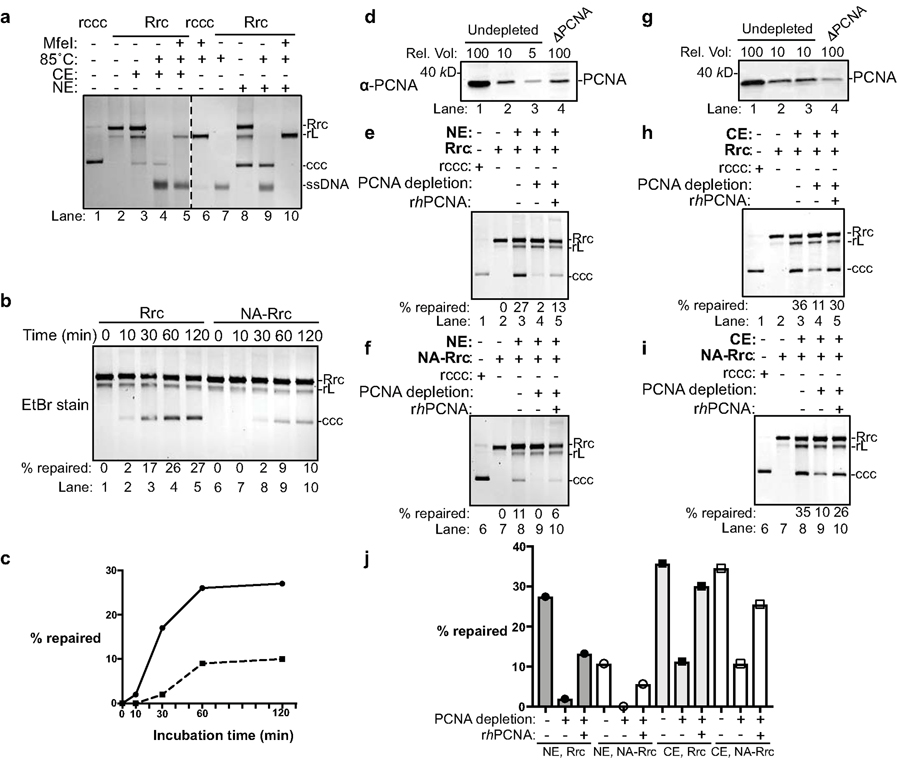
Fig. 3 |. PCNA is required for cccDNA formation in human cell extracts.
(a) Cytoplasmic and nuclear extracts from hNTCP-expressing HepG2 cells fully support cccDNA formation. Repair of RrcDNA was carried out and analyzed as in Fig. 2a, except the incubation temperature was 37°C instead of 30°C. Dashed line indicates removal of superfluous lanes. (b) Time course assay showing the kinetics of cccDNA formation from both RrcDNA (lanes 1–5) and NA-RrcDNA complex (lanes 6–10) in nuclear extracts. Experiments were performed as in (a), except that reactions were terminated at indicated time points. Efficiency of cccDNA formation was calculated as in Fig. 2c and indicated in row ‘% repaired’. (c) The efficiency of cccDNA formation from (b) is plotted against incubation time. (d) Immuno-depletion of more than 90% of PCNA from human nuclear extracts. Un-depleted and PCNA-depleted extracts were analyzed by western blotting using an anti-PCNA antibody. A relative volume (rel. vol) of 100 corresponds to 0.5 μl extract. Depletion of PCNA drastically reduced repair efficiency of RrcDNA (e) and NA-RrcDNA (f) in human nuclear extracts. Untreated recombinant cccDNA (Rccc) and Rrc were used as control (lanes 1, 2). Lanes 3 and 8, mock-depleted extract with mouse IgG; lanes 4 and 9, PCNA-depleted extract from (d) was used; lanes 5 and 10, addition of recombinant human PCNA (rhPCNA, shown in Fig. 4a) to PCNA-depleted extract restored cccDNA repair. (g–i) same to (d–f), except that human cytoplasmic extracts were used. (j) The efficiency of cccDNA formation from (e–f, g–i) is plotted. All data shown were repeated twice independently.