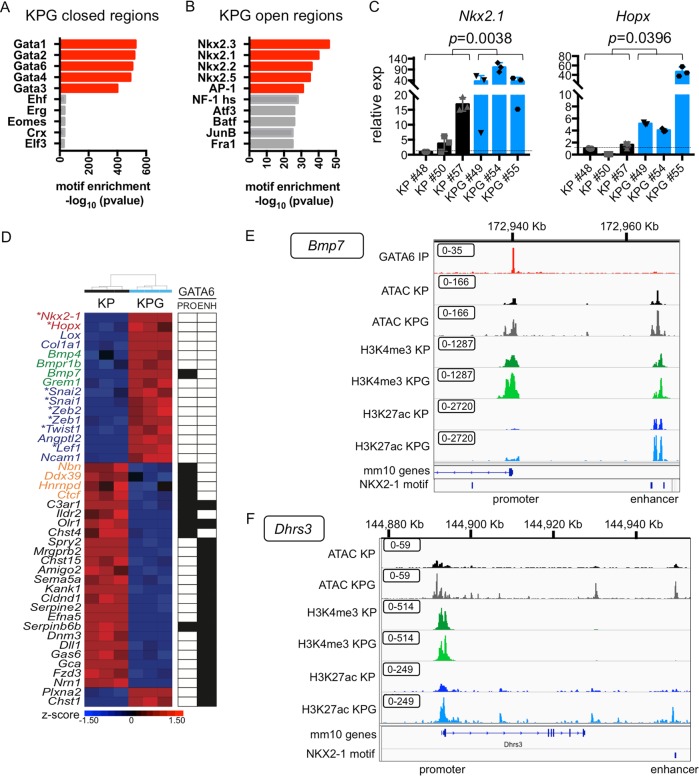
Fig. 5. GATA6 deficiency reprograms distal enhancers that are enriched for NKX2-1 motifs.
a TF motif analysis was performed for regions with newly closed chromatin in KPG cells. Log2 fold change < −1. P adjusted < 0.05. b TF motif analysis was performed for regions with newly open chromatin in KPG cells. Log2 fold change > 1. P adjusted < 0.05. c qRT-PCR of the indicated genes in cell lines from Fig. 3f (n = 3 biological replicates). Gene expression was normalized to b-actin. SEM is plotted and P value was calculated by Welch’s t-test. d Heatmap of representative genes differentially expressed in KPG versus KP cells (P adjusted < 0.05) clustered by Pearson average (z-score depicted). Genes involved in alveolar differentiation (red), BMP signaling (green), EMT (blue), and E2F targets (orange) are annotated. *TFs. For each gene, GATA6 binding (black box) in KP cells was indicated for their promoter (PRO) or enhancer (ENH). e IGV track for the upstream TSS of Bmp7 (Chr 2 172,918,000–172,970,000 of the mm10 genome) with overlapping GATA6 binding peaks at the promoter and an NKX2-1 motif overlapping a distal enhancer. f IGV track at TSS of Dhrs3 (Chr 4 144,878,000–144,950,000 of the mm10 genome) with an NKX2-1 motif overlapping a distal enhancer. Annotation of the tracks: KP GATA6 IP peaks (red), KP ATAC peaks (black), KPG ATAC peaks (gray), KP H3K4me3 peaks (dark green), KPG H3K4me3 peaks (light green), KP H3K27ac peaks (dark blue), KPG H3K27ac (light blue). Boxed values = data range for each track.