Abstract
Owing to the complexity of interacting molecular networks on the cell surface, integrin-associated tetraspanin CD151 remains controversial regarding its clinical importance and functional impact in prostate cancer. The current study evaluated dynamics and clinical importance of CD151 expression and its function in prostate cancer by IHC analysis of two independent patient cohorts (n=80, 181), bioinformatic interrogation of the TCGA database, and evaluation of gene knockdown effect at the cellular level. Our data showed that aside from high mRNA expression, CD151 was primarily localized to intercellular junctions at the plasma membrane in normal prostate glands or benign tissues, regardless of nature of antibodies used. By contrast, in primary tumors from patients with metastatic disease, CD151 was largely localized in the cytosol. Furthermore, the level of the cell-cell junction-linked CD151 was inversely associated with Gleason grade and tumor stage (P<0.001 for both). The portion of primary tumors expressing junctional CD151 was also three-fold less in the metastatic patient population than its counterpart (P<0.001). In line with these observations, CD151 and its associated α3β1 or α6β4 integrin inversely correlated with androgen receptor (AR) at the mRNA level (Spearman coefficient: -0.44, -0.48 and -0.42) in the TCGA cohort. Expression of these adhesion molecules also correlated with DNA methylation in their promoters (Spearman coefficient: -0.37, -0.71 and -0.82). Combined, these data suggest that CD151 and associated integrins are linked to tumor metastasis through AR and the epigenetic program. Meanwhile, CD151 knockdown in E-cadherin-positive tumor cells led to increased cell proliferation and induction of the epithelial-mesenchymal transition (EMT)-like phenotype. Given the strong RGD-binding integrin dependence of EMT-featured tumor cells, we examined focal adhesion kinase (FAK), their key signaling effector, in the above patient cohorts. In contrast to CD151, FAK exhibited positive correlation with tumor grade and stage as well as AR and p53 inactivation at either mRNA, protein or genomic level. Taken together, our results suggest that CD151 represses prostate cancer by antagonizing cell proliferation, EMT and the signaling of RGD-binding integrins. Since this anti-tumorigenic role is prone to the AR-mediated transcriptional and epigenetic regulation, CD151 and possibly α3β1 and α6β4 integrins are of potential biomarkers for metastatic prostate cancer.
Keywords: Prostate cancer, tetraspanin, CD151, metastasis, EMT, integrins
Introduction
Despite advances in targeted therapies, prostate cancer remains one of the deadliest diseases among men world-wide [1]. Extensive genome-wide studies have revealed that prostate tumors undergo a wide range of aberrant molecular changes at genetic, somatic, and epigenetic levels throughout disease onset and progression [2-5]. Thus, there is still an unmet need for precisely defining molecular regulators of prostate cancer to facilitate development of personalized targeted therapies.
Multiple members of the tetraspanin family, including CD9, CD81, CD82, TSPAN1 and CD151, exhibit aberrant expression during prostate tumor progression and metastasis [6-9]. Of particular interest is that CD82 and CD9 are strongly implicated as suppressors for metastatic prostate cancer [10,11]. This notion largely stems from studies comparing expression of these tetraspanins between primary and metastatic tumors [12-14]. Mechanistically, these integral membrane proteins appear to modulate cytoskeletal remodeling, intracellular signaling, and associated cell motility and invasion through their molecular partners such as EWI-2 or laminin-binding (LB) integrins, and particularly the α6β1 integrin [10,15,16].
In contrast to CD9 and CD82, tetraspanin CD151 is largely regarded as a driver of prostate cancer malignancy in the majority of published studies [8]. In an early IHC-based study, CD151 expression in prostate tumors was found to correlate with Gleason grade and poor clinical outcomes [17]. Recent experimental analysis also showed that CD151 deletion led to a decrease in lung-oriented tumor metastasis in the TRAMP model, an atypical mouse model for recapitulating development of neuroendocrine prostate cancer [18]. This pro-malignant function appears independent of the laminin-binding (LB) integrin-mediated signaling [19]. However, there is also growing evidence that CD151 is a multifaceted mediator of human cancer development and progression, particularly for epithelial-origin tumors such as bladder, breast, colon, endometrial and ovarian cancers [20-26]. This notion is supported by the ability of CD151 to stabilize cell-cell adhesion and to sequester the signaling of the HIPPO or Wnt pathways [27-29]. Given these conflicting observations on CD151 in prostate cancer and other epithelial-origin carcinomas, there is a particular need for more studies investigating its expression across different stages of carcinogenesis and clinical progression.
Here we critically examined the link between CD151 expression and prostate cancer malignancy. To gain unbiased evidence, our IHC analysis was conducted with two distinct monoclonal antibodies and two independent patient cohorts. Differences in CD151 expression between tumor stages and grades were systematically analyzed with respect to staining intensity and subcellular localization. Furthermore, both TCGA and local patient cohorts were investigated regarding the association between CD151/α3β1/α6β4 integrin complexes and AR status, as well as the RGD-binding integrin/FAK-dependent signaling. Finally, the functional role of CD151 in E-cadherin-positive prostate tumor cells was assessed with multiple hairpin shRNA or RNAi oligos. Overall, our data supports a context-dependent role of CD151 in prostate cancer and challenges the current unidirectional view of CD151 function and signaling during disease progression.
Materials and methods
Cell lines and antibodies
Human prostate cell lines, including BPH, 22RV1 and PC-3, were obtained from ATCC (Gaithersburg, MD). Cells were cultured in RPMI 1604 or DMEM (Invitrogen) medium supplemented with 5-10% FBS (Sigma-Aldrich, St; Louis, Mo) at 37°C and 5% CO2. All cell lines were periodically examined by PCR for Mycoplasma contamination [30]. The sources of antibodies and chemical inhibitors are described in a prior study [31]. The CD151 antibodies used for IHC staining included NCL-CD151 from Leica (Buffalo Grove, IL) and 11G5A from Abcam (Cambridge, MA). The 5C11 and 1A5 monoclonal antibodies used for FACS analysis and immunoblotting were raised in house or purchased from BioLegend (San Diego, CA). The IHC-oriented anti-α3 integrin antibody was obtained from Sigma (St. Paul, MS). The D23 clone of rabbit anti-α3 integrin cytoplasmic tail was generated in-house [20]. The antibodies against pY416-Src or pY397 and total FAK were obtained from Cell Signaling Technology (Danver, MA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. The chemical inhibitors, including VS-6063, Dasatinib and ICG001, were purchased from Sellechem (Houston, TX).
Transfection and expression of open reading frame constructs siRNA oligos and shRNA
The siRNA oligos of CD151 and α3 integrin were obtained from Cell Signaling Technology and Dharmacon (Boulder, CO). Cell transfection with siRNA oligos or DNA constructs was conducted using Lipofectamine 2000 (ThermoFisher, Waltham, MA). The stable knockdown of CD151 was carried out using pLKO lentiviral infection [31] or GIPZ vector from Dharmacon (Denver, CO).
Analyses of cellular functions and signaling
Cell viability and proliferation were analyzed with the MTT assay and counting of cell number microscopically as previously described [31]. For immunoblotting analysis of CD151 expression, tumor cells were lysed in RIPA buffer supplemented with protease inhibitors, followed by detection of CD151 proteins in 1A5 monoclonal antibody (BioLegend) under non-reducing and non-boiled condition and the Supersignal Chemoluminescence kit (Thermo-Fisher, Waltham, MA).
Construction and IHC analysis of TMAs of localized prostate tumors
Paraffin-embedded tissue blocks were selectively cored from prostatectomy specimens from 165 patients with prostate cancer and 34 patients with benign tumors for constructing a tissue microarray (TMA) through the Markey Cancer Center Biospecimen and Tissue Procurement Shared Resource, University of Kentucky (Lexington, Kentucky). Approval for use of human prostate tissue was obtained from the University of Kentucky Institutional Review Board. Tissue cores (2 mm) containing cancerous tissues, adjacent benign epithelial tissues, or benign prostatic hyperplasia (BPH) was used for the construction of tissue microarrays with duplicate cores from each patient. A single Gleason Grade was designated to each core prospectively during TMA construction by a single reviewer (PJH). Pathologic stages were obtained from retrospective review of pathology reports. Clinical outcomes data were obtained from retrospective chart review and Kentucky Cancer Registry outcomes data. Additionally, a commercial TMA harboring prostate tumors was obtained from US Biomax, Inc (Derwood, MD).
TMA slides were deparaffinized in a 60°C oven for 1 hour and heat-induced epitope retrieval was performed using a digital decloaking chamber (BioCare Medical, Concord, CA) for 20 minutes at 110°C in 1.0 mM EDTA (pH=9.0). Immunoreactivity was determined based on “Quick Score” calculated by multiplying staining intensity (on a scale of 1-3) by percentage of tumor cells with positive immunoreactivity by two blinded reviewers. The subcellular localization of CD151 proteins was stratified as “diffuse” or “cell-cell junction” pattern, which were defined as equal distribution of cytosolic and plasma membrane staining or predominant plasma membrane staining concentrated at areas of intercellular interactions.
Bioinformatic and statistical analyses
The Cancer Genome Atlas (TCGA) prostate cancer dataset (Cell, 2015) was interrogated for mRNA expression of relevant genes in human prostate tumors. The results shown are based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. The Chi-Square test and two-tailed T-tests with Welch’s correction for unequal sample size were used to evaluate differences in subcellular localization of CD151 or FAK proteins in primary tumors and associated clinical outcomes. The statistical significance of differences was set at P<0.05.
Results
Expression of CD151 in primary tumors from independent patient cohorts
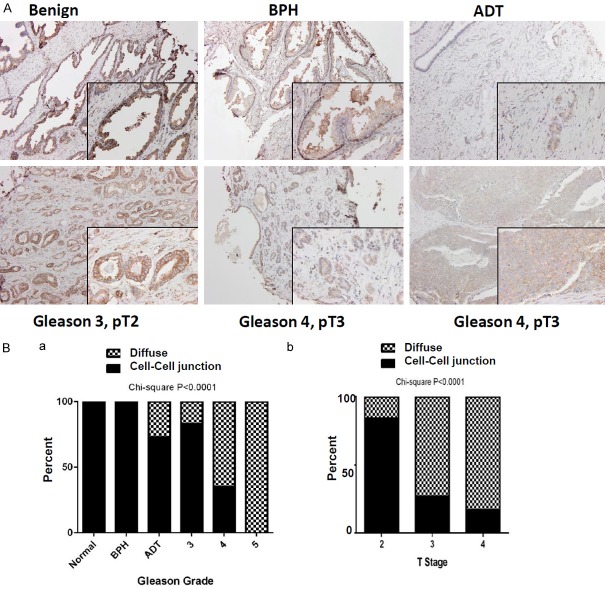
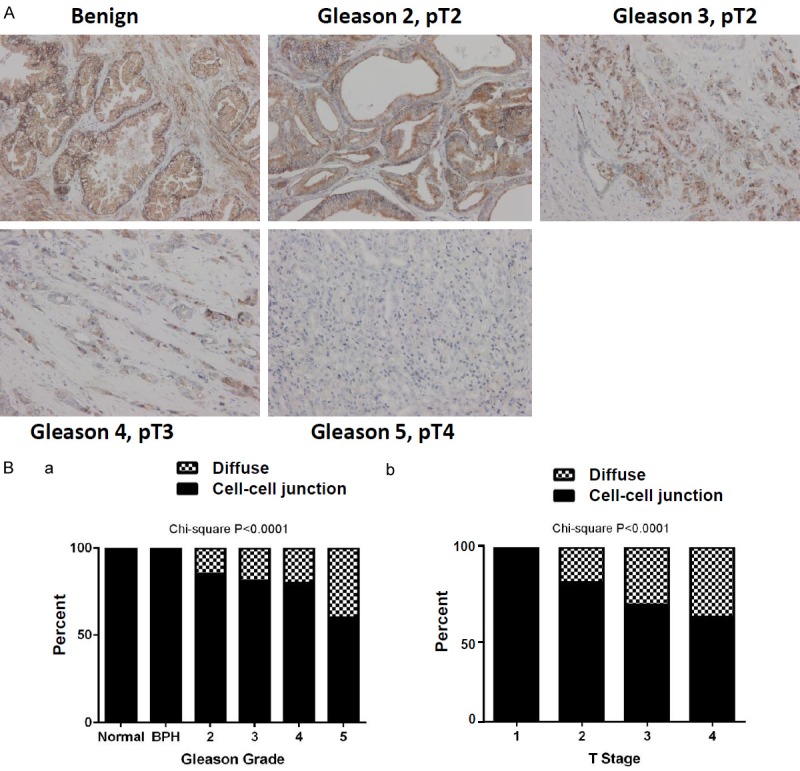
To evaluate the clinical link between CD151 and human prostate cancer, NCL-CD151, a widely used CD151-specific monoclonal antibody [20,32,33], was adopted for IHC analysis of a TMA harboring 80 individual patient cores. As shown in Figure 1 and Table 1, CD151 was strongly localized to cell-cell junctions in non-neoplastic tissues. The intensity of this intercellular distribution exhibited a decrease with increasing tumor stage and grade (P<0.001 for both). By comparison, the cytoplasmic fraction of CD151 protein was elevated in tumors at advanced stages. Furthermore, CD151 expression was independently examined with an independent TMA, which was constructed at the University of Kentucky with 181 unique prostate tumor cores. The 11G5A antibody (Abcam) was utilized for this TMA analysis since it recognizes the total pool of CD151 in tumor cells, irrespective of engagement with LB integrins or other partners [34]. As shown in Figure 2 and Table 2, CD151 was similarly localized to intercellular junctions at the plasma membrane in benign tissues, but this specificity decreased with advanced stage and grade tumors. It was also accompanied by increased cytosolic staining in tumor tissues. In line with these observations, CD151 is strongly expressed at the mRNA level in normal prostate glands or benign tissues, according to our interrogation of the Oncoming database (data not shown). Collectively, our two parallel analyses reveal an inverse association between the intercellular expression of CD151 at cell-cell contact points and tumor progression in prostate cancer, regardless of antibody epitope.
Figure 1.
Profiling CD151 expression in a commercial prostate cancer TMA patient cohort (N=80). Tissue was stained for CD-151 with the 11G5a monoclonal antibody (Abcam). A. Representative image of diverse CD151 staining patterns in primary tumors (100×). B. Subcellular localization of CD151 stratified by Gleason grade (a) and tumor stage (b). IHC imaging are representative of staining patterns and intensity by stage/grade. Percentages of diffused (filled bar) or cell-cell adhesion (hashed bar) were plotted against each of Gleason grades and tumor stages. Differences between groups were assessed using the Chi-Square analysis and p-values were indicated.
Table 1.
The TMA/IHC analysis of association between CD151 and clinical paramaters in the Biomax prostate cancer patient cohort (N=80)
| Variable (N) | CD151 staining | ||
|---|---|---|---|
|
| |||
| Ave Quick Score | STDP | p Value | |
| Grade | |||
| Control (10) | 255 | 79.9 | |
| BPH (20) | 231.3 | 69.8 | |
| Gleason 2 (9) | 172.2 | 115.9 | <0.0001 |
| Gleason 3 (20) | 127.4 | 99.7 | |
| Gleason 4 (11) | 103.2 | 96.6 | |
| Gleason 5 (10) | 52 | 36.8 | |
| Stage (N) | |||
| T1 (1) | 250 | 57.7 | |
| T2 (25) | 138.3 | 100.4 | <0.0001 |
| T3 (18) | 98.7 | 97.3 | |
| T4 (5) | 34 | 31.5 | |
| Lymph Node Metastasis (N) | |||
| Yes (17) | 77.5 | 72.5 | <0.0001 |
| No (33) | 134.4 | 106.8 | |
| Visceral/Bone Metastasis (N) | |||
| Yes (12) | 82.5 | 80.5 | <0.0006 |
| No (34) | 133.4 | 104.7 | |
Figure 2.
Profiling CD151 expression in a local prostate cancer TMA patient cohort (N=181). Tissue was stained with the NCL-CD151 monoclonal antibody. A. Representative image of diverse CD151 staining patterns in primary tumors (100×, 200× insert). B. Subcellular localization of CD151 stratified by Gleason grade and tumor stage. IHC imaging are representative of staining patterns and intensity by stage/grade. Percentages of diffused (filled bar) or cell-cell adhesion (hashed bar) were plotted against each of Gleason grades and tumor stages. Differences between groups were assessed using the Chi-Square analysis and p-values were indicated.
Table 2.
The TMA/IHC analysis of association between CD151 and clinical parameters in a local prostate patient cohort (N=181)
| Variable (N) | CD151 staining | ||
|---|---|---|---|
|
| |||
| Ave Quick Score | STD | p Value | |
| Grade | |||
| Control (21) | 287.9 | 28.3 | <0.0001 |
| BPH (7) | 262.1 | 48.7 | |
| ADT (11) | 222.7 | 85.2 | |
| Gleason 3 (69) | 242.5 | 67.7 | |
| Gleason 4 (60) | 214.6 | 83.1 | |
| Gleason 5 (13) | 242.2 | 83.7 | |
| Stage (N) | 0.1002 | ||
| T2 (80) | 246.7 | 65.6 | |
| T3 (67) | 210.3 | 85.3 | |
| T4 (6) | 246.3 | 50.4 | |
| Lymph Node Metastasis (N) | 0.9254 | ||
| Yes (7) | 229.2 | 75.3 | |
| No (146) | 230.8 | 77.5 | |
| Recurrence (N) | 0.4014 | ||
| None (111) | 229.4 | 91.9 | |
| Local (23) | 242.4 | 72.6 | |
| Distant (8) | 240.9 | 77.1 | |
| Disease-Specific Mortality (N) | 0.8633 | ||
| Survival (125) | 228.3 | 79.8 | |
| Death (9) | 231.1 | 83.9 | |
The clinical link between CD151 and metastatic progression
We subsequently examined the clinical link between CD151 and prostate tumor metastasis. The portion of diffuse staining of CD151 was significantly higher in primary tumors from pT stage- and grade-matched patients with known lymph node (pN+) or distant metastatic disease compared to those with organ-confined diseases in the Bio-max cohort (Figure 3A, P<0.0001 and <0.006, respectively) or the local patient cohort (Figure 3B, P<0.0001 for both). In contrast, the cell-cell junctional staining of CD151 exhibited a concomitant decrease in the group of patients with local or distant metastases in both cohorts. Combined, these data demonstrate that CD151 expression is altered in advanced prostate cancer, characterized by a marked shift from cell-cell junctional pattern towards diffuse and cytosolic localization.
Figure 3.
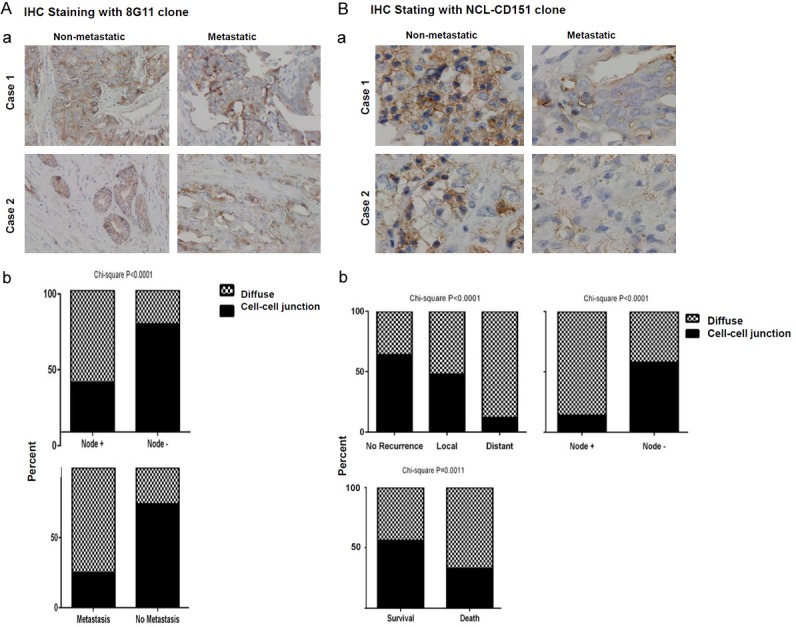
Differential expression of CD151 in primary tumors from patients with metastatic or non-metastatic disease. A, B. Representative images of primary tumors from patients with distant metastasis and organ-confined diseases are depicted for comparison. Primary tumors were subjected to IHC staining with 11G5a or CD151-NCL monoclonal antibodies. Differences between groups were assessed by Chi-Square analysis. p-values were indicated. Percentages of diffused (filled bar) or cell-cell adhesion (hashed bar) in the cohort were calculated and plotted for each of patient groups without disease recurrence or with node or distant metastasis.
Furthermore, we evaluated the link between CD151 and prostate cancer malignancy at the genomic level by interrogating the TCGA dataset (Cell, 2015). As shown in Figure 4A, there was lack of apparent association between CD151 and Gleason grade at the mRNA level, in contrast to its protein expression (Figures 1, 2). The expression of CD151 mRNA, however, appeared inversely associated with mRNA expression of AR, a key oncogenic driver of prostate carcinogenesis (Figure 4Ab). In line with this observation, there was a strong inverse association between CD151 and degree of methylation of its promoter region (Figure 4Ac). Meanwhile, CD151 was positively associated with expression of key adhesion molecules involved in the cell-cell junction, including P-cadherin, E-cadherin and α-catenin (Figure 4Ad). Similar analyses also revealed a close link for CD151-associated α3β1 and α6β4 integrins, but not E-cadherin (Figure 4B-D). Collectively, these data suggest that the diminished expression of CD151 at cell-cell contact sites attributed to the AR-mediated transcriptional suppression of CD151 and associated α3β1 and α6β4 integrins at the epigenetic level, corroborating its inverse association with advanced disease (Figures 1, 2 and 3).
Figure 4.
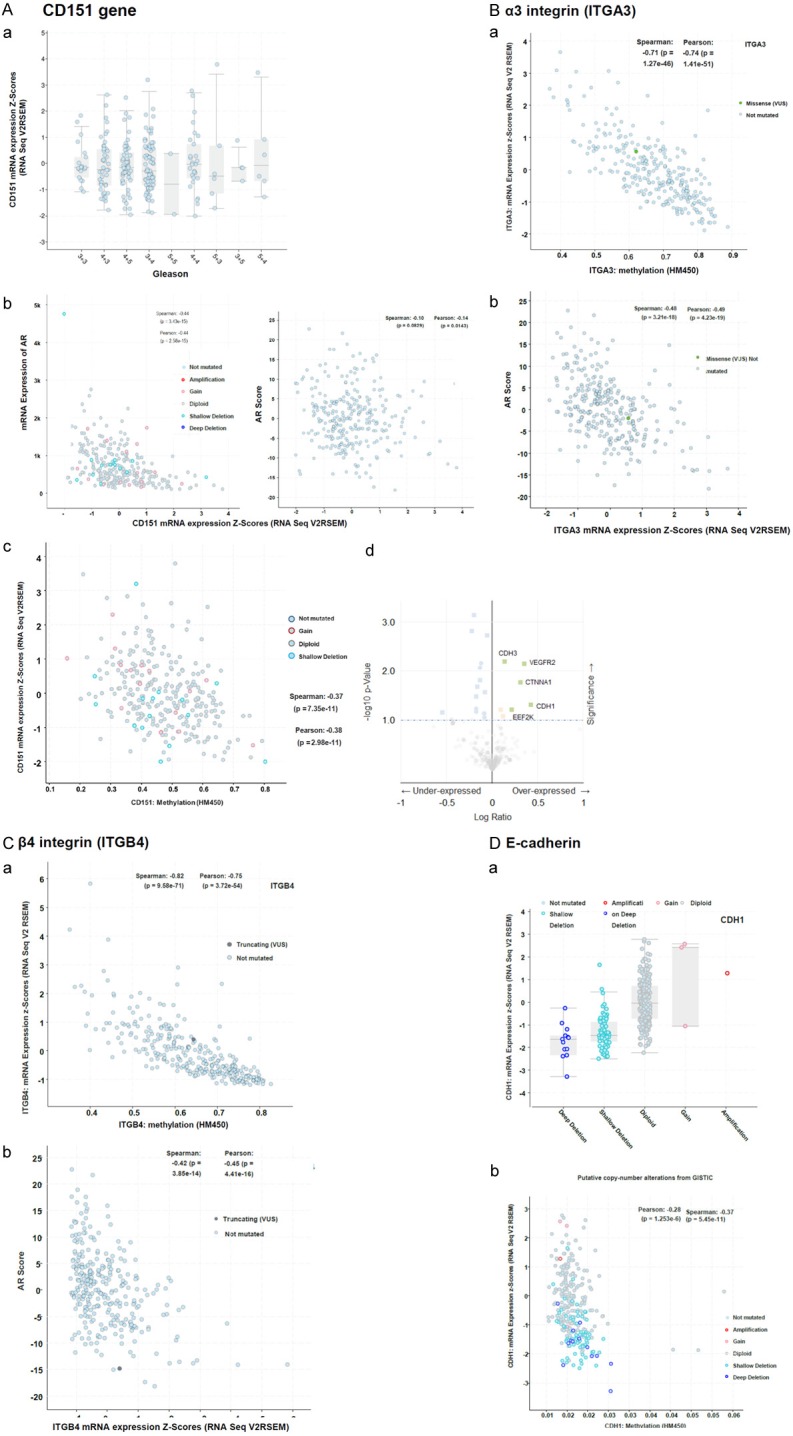
A link between epigenetic and AR regulation and expression of CD151 and associated α3β1 and α6β4 integrins in prostate tumors at mRNA level. The TCGA cohort (n=333, Cell 2015) was interrogated regarding: A. (a) the association between CD151 mRNA expression and Gleason grade; (b) A genomic link between CDd151 and AR. (c) Plots of CD151 mRNA expression and state of methylation of its promoter region. (d) Volcano blot of the list of genes being co-overexpressed or underexpressed with CD151 at mRNA level. B-D. The association between mRNA expression of α3β1 and α6β4 integrins and E-cadherin and AR or state of methylation of its promoter region (a, b). The Spearman and Pearson correlation coefficients were shown.
Effect of CD151 downregulation on tumor cell proliferation
We investigated functional consequences of decreased CD151 expression in advanced prostate tumors. As shown in Figure 5, attenuating CD151 expression via RNAi oligo or lentiviral-mediated expression of shRNAs markedly increased cell growth in an immortalized, E-cadherin-positive BPH cell line. However, downregulating α3 integrin only led to a moderate suppression of cell growth. A similar trend was observed for the metastatic, E-cadherin-positive 22RV1 prostate cancer cell line upon CD151 downregulation via expression of multiple hairpin shRNAs. Furthermore, tumor cells underwent an EMT-like morphological change in both cell lines in the absence of CD151 (Figures 5Ac, 5Bb, S1). These data demonstrate that CD151 represses tumor cell growth via promoting cell-cell contact and/or counteracting the EMT-like program.
Figure 5.
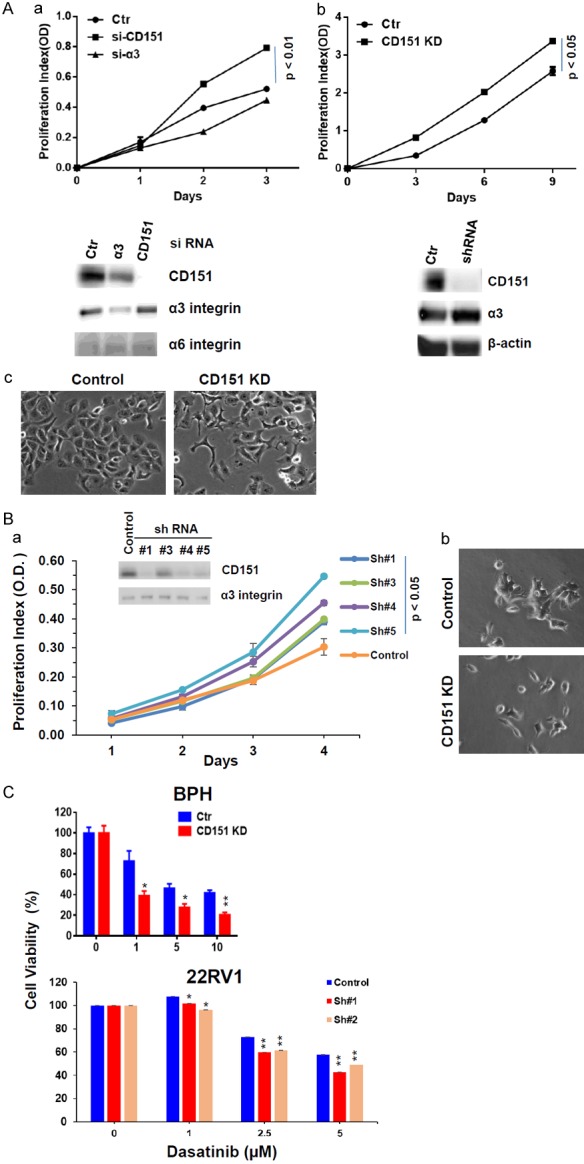
Effect of CD151 downregulation on proliferation and the EMT-like phenotype in cultured prostate tumor cells. A, B. Tumor cells were subjected to CD151 known via siRNA oligos or shRNA, followed by analysis of cell proliferation and morphologies. Degree of CD151 knockdown was determined by immunoblotting. A. BPH line; B. 22RV1 line. C. Modulation of tumor cell sensitivity to Dasatinib. Tumor cells with or without stable knockdown of CD151 were treated with indicated inhibitors for 72 h, followed by analyses of cell viability by MTT assay and paired t-test analysis. *: p value <0.05; **: p value <0.01.
The clinical association between FAK and prostate cancer aggressiveness
Based on the association between CD151 expression and advanced prostate cancer, we next investigated its role in intracellular signaling. Upon CD151 downregulation, tumor cells became more sensitive to inhibition of the RGD-binding integrin (α5β1 or αvβ3)-associated signaling through c-Src, which is known to promote the maintenance of E-cadherin/β-catenin complexes, as indicated by a decreased cell viability under escalating doses of its chemical inhibitor, Dasatinib (Figure 5C).
Since EMT induction is known to promote tumor cell dependence towards the RGD-binding integrin/FAK signaling axis, we examined the clinical relevance of this axis to gain additional evidence on CD151 function in this disease. As show in Figure 6A, the expression of FAK in human prostate cancer specimens was investigated. Expression of FAK increased with Gleason grade (P<.0001), pathologic stage (P<.0001), and prostate cancer-specific mortality (P<.0001), according to IHC analysis of the local patient cohort (Figure 6A and Table 3). Additionally, the average ratio of CD151: FAK staining in tumor tissue was 1.3 in Gleason 5 tumors, 1.7 in Gleason 4 tumors, 2.3 in Gleason 3 tumors, and 4.3 in non-neoplastic tissue. FAK expression also declined in tumors from patients managed with neoadjuvant androgen deprivation therapy (ADT, Figure 6 and Table 3). However, the ratio of CD151 versus FAK staining in ADT-treated patient tumors was still low (1.1).
Figure 6.
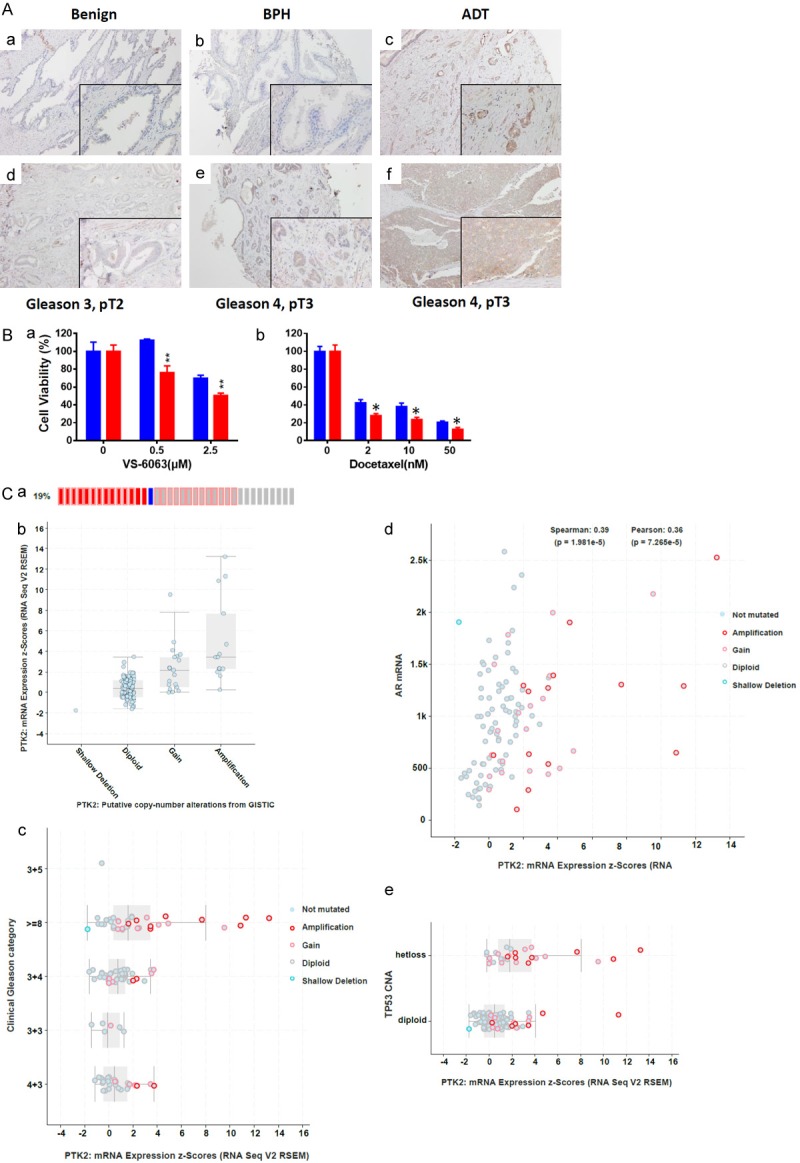
Reprehensive image of FAK staining in human prostate tumors. A. TMA from the local prostate cancer patient cohort was subjected to IHC analysis with an FAK-specific antibody. a-f. FAK staining in tumors with benign feature or varying in Gleason grade or stage. Scale: 100×, 200× insert. B. MTT analysis of CD151 knockdown on tumor cell sensitivity to FAK inhibitor (VS-6063) or chemotherapeutic agent (Docetaxel). BPH Tumor cells with or without stable knockdown of CD151 were treated with indicated agents for 72 h, followed by analyses of cell viability by MTT assay and paired t-test analysis. *: p value <0.05; **: p value <0.01. C. FAK deregulation at genomic and mRNA levels and association with oncogenic drivers in the TCGA prostate cancer patient cohort (Cell, 2015). a, b. Association between mRNA expression of FAK and gene copy number. CD151 mRNA expression and Gleason grade. c-e. Plots of FAK mRNA expression and tumor Gleason grade, AR and p53.
Table 3.
The TMA/IHC analysis of association between FAK, CD151 and clinical parameters in a local prostate patient cohort (N=181)
| Variable | FAK staining | |||
|---|---|---|---|---|
|
| ||||
| Average Score | STD | Value | CD151/FAK | |
| Grade (N) | ||||
| Control (21) | 66.6 | 34.9 | <0.0001 | 4.3 |
| BPH (7) | 61.8 | 30.4 | 4.2 | |
| ADT (11) | 202.3 | 85 | 1.1 | |
| Gleason 3 (69) | 99.1 | 61.4 | 2.4 | |
| Gleason 4 (60) | 127.4 | 68.7 | 1.7 | |
| Gleason 5 (13) | 188.1 | 86.9 | 1.3 | |
| Stage (N) | <-.0001 | |||
| T2 (80) | 104.7 | 66.1 | 2.4 | |
| T3 (67) | 142.8 | 78 | 1.5 | |
| T4 (6) | 200 | 85.3 | 1.2 | |
| Lymph node metastasis (N) | <0.1587 | |||
| Yes (7) | 163.8 | 91.4 | 1.4 | |
| No (146) | 123.7 | 74.8 | 1.9 | |
| Recurrence (N) | <0.2716 | |||
| None (111) | 122.1 | 73.6 | 1.9 | |
| Local (23) | 116.3 | 58.1 | 2.1 | |
| Distant (8) | 158.6 | 104.6 | 1.5 | |
| Disease-specific mortality (N) | <0.0001 | |||
| Survival (125) | 116.9 | 68.5 | 2.0 | |
| Death (9) | 239.4 | 75.2 | ||
Furthermore, we found the CD151 downregulation in BPH cell line increased tumor cell sensitivity to VS-6063, an inhibitor of active FAK, as well as docetaxel (Figure 6B). In comparison, there was lack of apparent effect for the inhibitors of the PI3K (GDC-2094) or Wnt pathway (ICG001) (data not shown). Collectively, these observations demonstrate an active contribution by the RGD-binding integrin-FAK signaling pathway towards advanced stage and grade prostate tumors, counteracting the role of CD151/α3β1 or α6β4 integrin complex in maintenance of cell-cell contact.
Discussion
The current study has evaluated the differences in CD151 expression and subcellular localization between tumor grades and stages across parallel prostate cancer patient cohorts. The data indicate that CD151 is abundantly localized to cell-cell junctions and on the cell surface in non-cancerous tissues, and this expression pattern diminishes in with escalating tumor Gleason grade and stage, and presence of nodal or distance metastases. Moreover, this clinical trend is positively associated with expression of E-cadherin, P-cadherin and α-catenin during prostate cancer progression, and is inversely linked to occurrence of AR expression and DNA methylation. Downregulating CD151 in vitro not only leads to increased tumor cell growth, but induces an EMT-like morphological change. Our study also reveals that in contrast to CD151, FAK, a key signaling effector of EMT-linked RGD-binding integrins, is frequently amplified or overexpressed in advanced-stage prostate carcinomas. Overall, our results suggest a suppressive role of CD151 in prostate cancer, challenging the current paradigm of its impact on prostate tumor metastasis and its utility for defining aggressiveness of the disease [9,18].
Clinical relevance of altered CD151 expression and localization
Our results demonstrate that CD151 is primarily expressed at cell-cell contact interfaces in normal prostate or benign tumors, and this pattern diminishes with increased tumor Gleason grade and or progression to advanced metastatic disease (Figures 1, 2 and 3). This correlates with the evidence that the majority of prostate tumors appear to stem from the oncogenic targeting of luminal epithelial cells in prostate glands. In this case, CD151 function is partially aligned with the E-cadherin/β-catenin complexes, besides laminin-biding α3β1 and α6β4 integrins. This argument is supported by our functional analysis CD151 downregulation and its concordance with E-cadherin in prostate tumors, as well as AR or epigenetic regulation of CD151 and associated integrins at transcriptional level (Figures 4, 5). Hence, our study supports a cancer type- and oncogenic context-associated role of CD151 and laminin-biding α3β1 and α6β4 integrins in human epithelial-origin cancers [8,20], rather than its subcellular localization on tumor cells [19]. It also argues for the role of CD151 as a biomarker in prostate cancer progression.
The IHC expression profile of CD151 exhibited in the current study is not entirely in agreement with some prior analyses [17,19,35]. One argument for this discrepancy is the variability of CD151 antibodies in recognizing different domains of CD151 proteins present on tumor cell surface [36]. Since use of two IHC application-tested antibodies fails to yield any staining of α3β1 integrin in in primary tumor tissues from our above cohorts (data not shown) [27,37], it remains an open question that CD151 proteins at the cell-cell junctions belong to the pool of the “integrin-free” molecules on the cell surface.
Molecular regulation of CD151 expression during prostate tumor growth and progression
A notable observation from the current study is the evidence of transcriptional and epigenetic regulation of CD151 in prostate cancer. CD151 is known to undergo post-transcriptional regulation, including co-expression with integrins and clathrin-coated pit-associated recycling [38,39]. Our data suggest that this regulation is likely tied to the action of AR, which is also known to drive prostate tumorigenesis and progression via epigenetic silencing of tumor suppressors. This is also consistent with a recent study of TSPAN1 in prostate cancer [40,41]. Reduced CD151 expression at cell-cell contact domains may result from the increased methylation of its promoters during tumor progression (Figure 4) [19]. In fact, the promoter region of the CD151 gene is highly enriched in CpG islands (data not shown). This molecular feature may provide another key basis for the anti-metastatic role of CD151 in prostate cancer.
Plasticity of CD151 function in prostate cancer
Besides an anti-tumorigenic role during prostate tumor development highlighted in the current study (Figures 1, 2, 3, 4 and 5), CD151 may be pro-tumorigenic in tumor cells with squamous carcinoma-like or post-EMT mesenchymal phenotype. In this case, CD151 levels on the cell surface are largely dependent on expression of the ECM-binding α3β1 or α6β4 integrins. By forming complexes, these molecules collaborate with RGD-binding integrins (e.g., α2β1, α5β1 or αvβ3) to drive tumor cell motility and invasion through intracellular signaling of the integrin/FAK axis. This scenario is consistent with our analysis with squamous carcinoma-like PC-3 cell line, where attenuating CD151 expression impaired tumor cell motility stimulated by the HGF/c-Met axis (data not shown). This evidence is also supported by high levels of endogenous laminin-5 in PC-3 cells, which promotes CD151-α6β4 complexes-dependent cellular growth and aggressive behaviors in an autocrine manner [42]. Hence, the role of CD151 in prostate cancer may vary with tumor stage or subtype/oncogenic context, besides its molecular partners on plasma membrane, similar to many other key components of cell adhesion networks in human epithelial-origin tumors [20,21].
In summary, our analysis of prostate cancer patient biopsies reveals a strong association between subcellular distribution and transcription of CD151 with advanced Gleason grade, stage, and metastatic potential. This anti-tumor function is inversely linked to the regulation by AR at mRNA or epigenetic level. Hence, our study provides evidence on a putative anti-malignant function of CD151 and associated α3β1 and α6β4 integrins, challenging the current paradigm of its driver role in prostate cancer progression.
Acknowledgements
The authors acknowledge the support of this work by the James F. Hardymon Endowment in Urology Research (PH and NK), a pilot project award from NIH COBRE grant (5 P20 GM121327-03) to XY, the Biospecimen Procurement & Translational Pathology Shared Resource Facility of the University of Kentucky Markey Cancer Center (P30CA177558), and National Natural Science Foundation of China (81572928, 81772978) and Jiangsu Provincial Special Program of Medical Science (E2017611) to JC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer. 2015;137:262–266. doi: 10.1002/ijc.28940. [DOI] [PubMed] [Google Scholar]
- 2.Russo JW, Balk SP. Initiation and evolution of early onset prostate cancer. Cancer Cell. 2018;34:874–876. doi: 10.1016/j.ccell.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, Heckmann D, Sidiropoulos N, Waszak SM, Hubschmann D, Urbanucci A, Girma EG, Kuryshev V, Klimczak LJ, Saini N, Stutz AM, Weichenhan D, Bottcher LM, Toth R, Hendriksen JD, Koop C, Lutsik P, Matzk S, Warnatz HJ, Amstislavskiy V, Feuerstein C, Raeder B, Bogatyrova O, Schmitz EM, Hube-Magg C, Kluth M, Huland H, Graefen M, Lawerenz C, Henry GH, Yamaguchi TN, Malewska A, Meiners J, Schilling D, Reisinger E, Eils R, Schlesner M, Strand DW, Bristow RG, Boutros PC, von Kalle C, Gordenin D, Sultmann H, Brors B, Sauter G, Plass C, Yaspo ML, Korbel JO, Schlomm T, Weischenfeldt J. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell. 2018;34:996–1011. e1018. doi: 10.1016/j.ccell.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin MA, Demichelis F. The genomics of prostate cancer: emerging understanding with technologic advances. Mod Pathol. 2018;31:S1–11. doi: 10.1038/modpathol.2017.166. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright MD, Moseley GW, van Spriel AB. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens. 2004;64:533–542. doi: 10.1111/j.1399-0039.2004.00321.x. [DOI] [PubMed] [Google Scholar]
- 7.Rubinstein E. The complexity of tetraspanins. Biochem Soc Trans. 2011;39:501–505. doi: 10.1042/BST0390501. [DOI] [PubMed] [Google Scholar]
- 8.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 9.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. doi: 10.1017/S1462399409001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003;63:2665–2674. [PubMed] [Google Scholar]
- 11.Ovalle S, Gutierrez-Lopez MD, Olma N, Turnay J, Lizarbe MA, Majano P, Molina-Jimenez F, Lopez-Cabrera M, Mo MY, Sanchez-Madrid F, Cabanas C. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int J Cancer. 2007;121:2140–52. doi: 10.1002/ijc.22902. [DOI] [PubMed] [Google Scholar]
- 12.Lijovic M, Somers G, Frauman AG. KAI1/CD82 protein expression in primary prostate cancer and in BPH associated with cancer. Cancer Detect Prev. 2002;26:69–77. doi: 10.1016/s0361-090x(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 13.Hashida H, Takabayashi A, Tokuhara T, Hattori N, Taki T, Hasegawa H, Satoh S, Kobayashi N, Yamaoka Y, Miyake M. Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer. 2003;89:158–167. doi: 10.1038/sj.bjc.6601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JC, Begin LR, Berube NG, Chevalier S, Aprikian AG, Gourdeau H, Chevrette M. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin Cancer Res. 2007;13:2354–2361. doi: 10.1158/1078-0432.CCR-06-1692. [DOI] [PubMed] [Google Scholar]
- 15.He B, Liu L, Cook GA, Grgurevich S, Jennings LK, Zhang XA. Tetraspanin CD82 attenuates cellular morphogenesis through down-regulating integrin alpha6-mediated cell adhesion. J Biol Chem. 2005;280:3346–3354. doi: 10.1074/jbc.M406680200. [DOI] [PubMed] [Google Scholar]
- 16.Murray NP, Aedo S, Fuentealba C, Reyes E. 10 year biochemical failure free survival of men with CD82 positive primary circulating prostate cells treated by radical prostatectomy. Asian Pac J Cancer Prev. 2018;19:1577–1583. doi: 10.22034/APJCP.2018.19.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang J, Lijovic M, Ashman LK, Kan K, Frauman AG. CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer Epidemiol Biomarkers Prev. 2004;13:1717–1721. [PubMed] [Google Scholar]
- 18.Copeland BT, Bowman MJ, Ashman LK. Genetic ablation of the tetraspanin CD151 reduces spontaneous metastatic spread of prostate cancer in the TRAMP model. Mol Cancer Res. 2013;11:95–105. doi: 10.1158/1541-7786.MCR-12-0468. [DOI] [PubMed] [Google Scholar]
- 19.Palmer TD, Martinez CH, Vasquez C, Hebron KE, Jones-Paris C, Arnold SA, Chan SM, Chalasani V, Gomez-Lemus JA, Williams AK, Chin JL, Giannico GA, Ketova T, Lewis JD, Zijlstra A. Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res. 2014;74:173–187. doi: 10.1158/0008-5472.CAN-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldwin LA, Hoff JT, Lefringhouse J, Zhang M, Jia C, Liu Z, Erfani S, Jin H, Xu M, She QB, van Nagell JR, Wang C, Chen L, Plattner R, Kaetzel DM, Luo J, Lu M, West D, Liu C, Ueland FR, Drapkin R, Zhou BP, Yang XH. CD151-alpha3beta1 integrin complexes suppress ovarian tumor growth by repressing slug-mediated EMT and canonical Wnt signaling. Oncotarget. 2014;5:12203–17. doi: 10.18632/oncotarget.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y, Deng X, Liu Z, Baldwin LA, Lefringhouse J, Zhang J, Hoff JT, Erfani SF, Rucker EB 3rd, O’Connor K, Liu C, Wu Y, Zhou BP, Yang XH. CD151 represses mammary gland development by maintaining the niches of progenitor cells. Cell Cycle. 2014;13:2707–22. doi: 10.4161/15384101.2015.945823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL. Does loss of CD151 expression promote the metastasis of hypoxic colon cancer cells? Clin Cancer Res. 2008;14:7969–7970. doi: 10.1158/1078-0432.CCR-08-2417. [DOI] [PubMed] [Google Scholar]
- 23.Chien CW, Lin SC, Lai YY, Lin BW, Lee JC, Tsai SJ. Regulation of CD151 by hypoxia controls cell adhesion and metastasis in colorectal cancer. Clin Cancer Res. 2008;14:8043–8051. doi: 10.1158/1078-0432.CCR-08-1651. [DOI] [PubMed] [Google Scholar]
- 24.Minner S, De Silva C, Rink M, Dahlem R, Chun F, Fisch M, Hoppner W, Wagner W, Bokemeyer C, Terracciano L, Simon R, Sauter G, Wilczak W. Reduced CD151 expression is related to advanced tumour stage in urothelial bladder cancer. Pathology. 2012;44:448–452. doi: 10.1097/PAT.0b013e32835576ee. [DOI] [PubMed] [Google Scholar]
- 25.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 26.Voss MA, Gordon N, Maloney S, Ganesan R, Ludeman L, McCarthy K, Gornall R, Schaller G, Wei W, Berditchevski F, Sundar S. Tetraspanin CD151 is a novel prognostic marker in poor outcome endometrial cancer. Br J Cancer. 2011;104:1611–1618. doi: 10.1038/bjc.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varzavand A, Hacker W, Ma D, Gibson-Corley K, Hawayek M, Tayh OJ, Brown JA, Henry MD, Stipp CS. alpha3beta1 integrin suppresses prostate cancer metastasis via regulation of the hippo pathway. Cancer Res. 2016;76:6577–6587. doi: 10.1158/0008-5472.CAN-16-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varzavand A, Drake JM, Svensson RU, Herndon ME, Zhou B, Henry MD, Stipp CS. Integrin alpha3beta1 regulates tumor cell responses to stromal cells and can function to suppress prostate cancer metastatic colonization. Clin Exp Metastasis. 2013;30:541–552. doi: 10.1007/s10585-012-9558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. alpha3beta1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J Cell Biol. 2003;163:1351–62. doi: 10.1083/jcb.200306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B, Lefringhouse J, Liu Z, West D, Baldwin LA, Ou C, Chen L, Napier D, Chaiswing L, Brewer LD, St Clair D, Thibault O, van Nagell JR, Zhou BP, Drapkin R, Huang JA, Lu ML, Ueland FR, Yang XH. Inhibition of the integrin/FAK signaling axis and c-Myc synergistically disrupts ovarian cancer malignancy. Oncogenesis. 2017;6:e295. doi: 10.1038/oncsis.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XH, Richardson AL, Torres-Arzayus MI, Zhou PC, Sharma C, Kazarov AR, Andzelm MM, Strominger JL, Brown M, Hemler ME. CD151 accelerates breast cancer by regulating alpha(6) integrin function, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romanska HM, Potemski P, Krakowska M, Mieszkowska M, Chaudhri S, Kordek R, Kubiak R, Speirs V, Hanby AM, Sadej R, Berditchevski F. Lack of CD151/integrin alpha3beta1 complex is predictive of poor outcome in node-negative lobular breast carcinoma: opposing roles of CD151 in invasive lobular and ductal breast cancers. Br J Cancer. 2015;113:1350–1357. doi: 10.1038/bjc.2015.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geary SM, Cambareri AC, Sincock PM, Fitter S, Ashman LK. Differential tissue expression of epitopes of the tetraspanin CD151 recognised by monoclonal antibodies. Tissue Antigens. 2001;58:141–53. doi: 10.1034/j.1399-0039.2001.580301.x. [DOI] [PubMed] [Google Scholar]
- 35.Schmelz M, Cress AE, Scott KM, Burger F, Cui H, Sallam K, McDaniel KM, Dalkin BL, Nagle RB. Different phenotypes in human prostate cancer: alpha6 or alpha3 integrin in cell-extracellular adhesion sites. Neoplasia. 2002;4:243–254. doi: 10.1038/sj.neo.7900223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kazarov AR, Yang X, Stipp CS, Sehgal B, Hemler ME. An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology. J Cell Biol. 2002;158:1299–309. doi: 10.1083/jcb.200204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JL, Winterwood N, DeMali KA, Stipp CS. Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J Cell Sci. 2009;122:2263–73. doi: 10.1242/jcs.045997. [DOI] [PubMed] [Google Scholar]
- 38.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, He B, Liu WM, Zhou D, Cox JV, Zhang XA. Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J Biol Chem. 2007;282:31631–42. doi: 10.1074/jbc.M701165200. [DOI] [PubMed] [Google Scholar]
- 40.Xu F, Gao Y, Wang Y, Pan J, Sha J, Shao X, Kang X, Qin J, You MJ, Huang Y, Dong B, Xue W. Decreased TSPAN1 promotes prostate cancer progression and is a marker for early biochemical recurrence after radical prostatectomy. Oncotarget. 2016;7:63294–63305. doi: 10.18632/oncotarget.11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munkley J, McClurg UL, Livermore KE, Ehrmann I, Knight B, McCullagh P, McGrath J, Crundwell M, Harries LW, Leung HY, Mills IG, Robson CN, Rajan P, Elliott DJ. The cancer-associated cell migration protein TSPAN1 is under control of androgens and its upregulation increases prostate cancer cell migration. Sci Rep. 2017;7:5249. doi: 10.1038/s41598-017-05489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell. 2009;20:2207–2217. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.