Fig. 5.
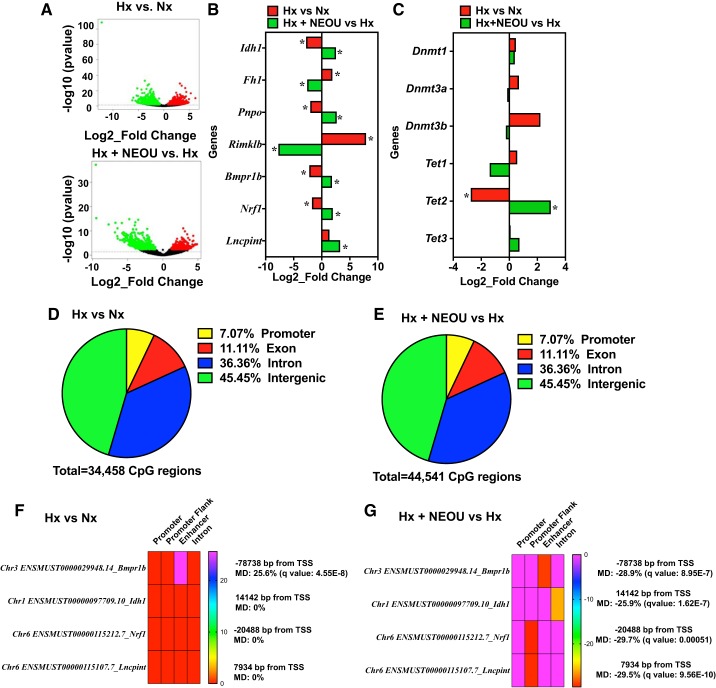
Pharmacological inhibition of glucose-6-phosphate dehydrogenase (G6PD) regulates the expression of noncoding and protein coding genes by controlling DNA methylation (epigenetic modification) in lungs of CYP2C44−/− hypoxic mice. Gene expression in lungs of normoxic (n = 3; male 2 and female 1), hypoxic (n = 3; male 2 and female 1), and hypoxic+NEOU (1.5 mg·kg−1·day−1; sc; n = 3; male 2 and female 1) was quantified by whole genome RNA-seq. A: volcano plots demonstrate several genes are upregulated and downregulated in the lungs of hypoxic vs. normoxic mice (top) and hypoxia+ N-ethyl-N′-[(3β,5α)-17-oxoandrostan-3-yl]urea (NEOU; 1.5·mg·kg−1·day−1; sc) vs. hypoxic mice (bottom). Red circles represent significantly upregulated genes, while green circles represent significantly downregulated genes between the groups. Black circles represent genes that were altered but not significantly. B: alterations in the expression of noncoding and protein coding [relevant to metabolic pathways and implicated in pulmonary hypertension (PH)] genes in lungs of hypoxia (Hx) vs. normoxia (Nx) mice, and hypoxia+NEOU (Hx+NEOU-treated for 1 wk) vs. hypoxia (Hx) mice is shown. C: expression of DNA (cytosine-5-)-methyltransferase 1, 3a, and 3b (Dnmt1, Dnmt3a, and Dnmt3b) and ten-eleven translocation methylcytosine dioxygenase 1, 2, and 3 (Tet1, Tet2, and Tet3) in lungs of hypoxic vs. normoxic mice and hypoxia+NEOU vs. hypoxic mice determined by RNAseq is shown. Change in gene expression is shown as Log2_fold change. *P < 0.05 (also see Table 1). DNA methylation in lungs of normoxic (n = 3; male 2 and female 1), hypoxic (n = 3; male 2 and female 1), and hypoxic+NEOU (1.5 mg·kg−1·day−1; sc; n = 3; male 2 and female 1) mice was determined by reduced representation bisulfite sequencing. D and E: pie graphs showing percentage of differentially methylated genes/regions (with a methylation difference threshold of 25% and a q value (SLIM) threshold of 0.05, calculated using the Chi-squared test) in lungs of hypoxia (Hx) vs. normoxia (Nx) and in hypoxia+NEOU (Hx+NEOU) vs. hypoxia (Hx), mice. A landscape map showing the DNA hypermethylation in same location on functional element of Bmpr1b in lung of hypoxia (Hx) vs. normoxia (Nx) (F), and DNA hypomethylation in functional elements of Bmpr1b, Idh1, Nrf1, and lncPint genes in lung of hypoxia+NEOU (Hx + NEOU) vs. Hx (G). Distance of functional elements from transcription start site (TSS), methylation difference (MD), and statistical significance are shown. Details of differential methylation analysis are given in the online supplement.