Keywords: comorbidity, COVID-19, fibrinolysis, plasmin(ogen), SARS-CoV-2
Abstract
Patients with hypertension, diabetes, coronary heart disease, cerebrovascular illness, chronic obstructive pulmonary disease, and kidney dysfunction have worse clinical outcomes when infected with SARS-CoV-2, for unknown reasons. The purpose of this review is to summarize the evidence for the existence of elevated plasmin(ogen) in COVID-19 patients with these comorbid conditions. Plasmin, and other proteases, may cleave a newly inserted furin site in the S protein of SARS-CoV-2, extracellularly, which increases its infectivity and virulence. Hyperfibrinolysis associated with plasmin leads to elevated D-dimer in severe patients. The plasmin(ogen) system may prove a promising therapeutic target for combating COVID-19.
Elevated plasmin(ogen) is a common feature in people with underlying medical conditions, including hypertension, diabetes, cardiovascular disease, cerebrovascular disease, and chronic renal illness, who are susceptible to SARS-CoV-2 infection.
Plasmin enhances the virulence and infectivity of SARS-CoV-2 virus by cleaving its spike proteins.
Extremely increased D-dimer in COVID-19 patients results from plasmin-associated hyperactive fibrinolysis.
D-dimer and viral load are independent risk factors of disease severity and mortality.
Antiproteases targeting plasmin(ogen) may be a promising approach to combat COVID-19.
I. INTRODUCTION
Patients with preexisting hypertension, diabetes, coronary heart disease, cerebrovascular disease, chronic obstructive pulmonary disease (COPD), and kidney dysfunction (comorbidities) have worse clinical outcomes when infected with SARS-CoV-2. The only treatment of COVID-19 is supportive (51), and registered clinical trials are ongoing. The mechanisms for high morbidity and mortality of patients with comorbidities are unknown. The existence of significantly increased fibrin degradation products (FDPs) and reduced platelets in severe COVID-19 patients is consistent with the presence of hyperfibrinolysis. This opinion is supported by the presence of hemorrhage in multiple organs and a positive correlation between fibrinolysis and mortality. Plasmin, a key player in fibrinolysis, enhances the virulence and pathogenicity of viruses containing a furin site in their envelope proteins, as is the case with the SARS-CoV-2. The purpose of this review is to summarize the clinical and preclinical evidence for the existence of elevated plasmin(ogen) in these comorbid conditions of COVID-19 and to highlight the importance of plasmin-induced proteolytic cleavage of the SARS-COV-2 S protein and fibrin in the development of COVID-19.
II. CLINICAL, PATHOLOGICAL, AND EPIDEMIOLOGICAL FEATURES OF COVID-19
A. Epidemiology
Coronavirus disease 2019 (COVID-19) is caused by a new β-coronavirus, SARS-CoV-2. Its epicenter was in Wuhan, China, and it has been spreading globally (103). As of March 31, 2020, there have been 858,955 cases worldwide resulting in 42,119 deaths.1 This far exceeds the total deaths caused by both severe acute respiratory syndrome (SARS) and Middle Eastern respiratory syndrome (MERS) (91). Persons older than 60 years with hypertension, diabetes, COPD, as well as cardiovascular, cerebrovascular, liver, kidney, and gastrointestinal diseases are more susceptible to the infection by SARS-CoV-2 and experience higher mortality when they develop COVID-19 (3, 80, 101). The contribution of malignant conditions is under debate due to the small number of patients (43, 92). In total, 19% of COVID-19 patients develop acute respiratory distress syndrome (ARDS) (Pao2/FiO2 <300 mmHg) within 24–48 h after onset of symptoms.
B. Viral Load
Virus clearance is associated with the severity and survival of COVID-19. Viral load in the respiratory tract peaks at day 5–6 after the onset of symptoms (104–107 copies/ml) (62), and viral RNA can be found in stool and sputum samples, bronchoalveolar lavage fluid (BAL), and lung epithelial cells. Patients older than 65 years generally have higher viral load lasting up to 14 days and may develop severe acute lung injury, requiring hospitalization in the intensive care unit (ICU) with poor outcome (62). In contrast, most younger patients have a much lower viral load that is undetectable within 1 week after onset (104). Furthermore, an association between viral load and the severity of COVID-19 has been reported (46).
C. Leading Causes of Death
COVID-19 patients admitted to ICU have higher mortality (38%) than non-ICU patients (4%) (31). The mortality of patients who develop ARDS is 49% (45). Many patients with COVID-19 develop multi-organ failure (MOF). The leading causes of deaths are ARDS, septic shock with MOF, hemorrhage/coagulopathy (disseminated intravascular coagulopathy, DIC), acute heart/liver/kidney injury, and secondary bacterial infections (97, 101). Elevated FDPs and D-dimers were detected predominantly in patients with severe disease (11, 24, 31, 43, 45, 79, 85). Multivariate regression further suggests that age and D-dimer levels (>1 mg/L) are two independent risk factors for mortality (89, 101). This in addition to other factors, such as compromised immune response, may contribute to the increased morbidity and mortality of patients with COVID-19 who are older than 60 years.
D. Pathology
The pathological features of COVID-19 resemble those of SARS and MERS. In the early stages of infection, puncture lung biopsies reveal the presence of pneumonia, edema, proteinaceous exudate with globules and focal hyperplasia of alveolar epithelial cells associated with patchy inflammatory infiltrates, and multinucleated giant cells (81). At the later stages, diffuse alveolar damage (DAD) is observed in addition to hemorrhage and some areas of interstitial fibrosis (48). Fibrotic clots and gelatinous mucus in the small airways and disseminated intravascular coagulation are also present (44, 93). Consistent with clinical observations, the lungs are the most injured organs, followed by moderate injury in the heart, liver, kidney, and brain. Systemic microthrombi in the circulatory system and hemorrhage in the affected organs result from noncoordinated responses between the coagulation and fibrinolysis systems. Although COVID-19 is characterized by hyperfibrinolysis, as evidenced by elevated levels of D-dimer, studies attempting to restore fibrinolytic function have not been reported. Elevated plasmin(ogen) in patients with preexisting conditions may be a mechanism contributing to enhanced susceptibility to SARS-CoV-2 infection and fatality.
III. CLEAVAGE OF SARS-CoV-2 S PROTEINS BY HOST FURIN AND PLASMIN
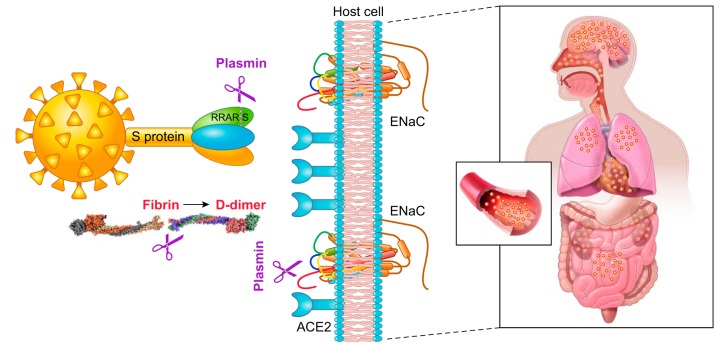
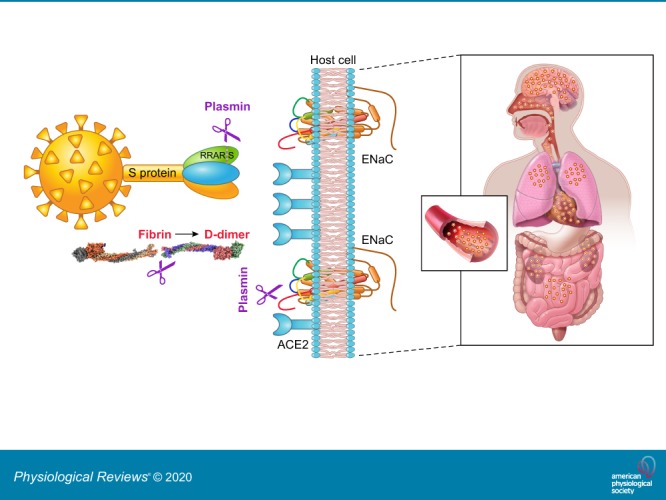
Sequencing of hundreds of SARS-CoV-2 virus isolates reveals a close relation to two bat-derived coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21. These coronavirus strains have a similar receptor-binding domain structure in the Spike (S) protein for host angiotensin converting enzyme 2 (ACE2) proteins (47, 90, 102). The S protein of SARS-CoV-2 bind to human ACE2 receptors with higher affinity than that of the SARS-CoV virus (88). This may be due to a furin-like cleavage site (682RRAR/S686) inserted in the S1/S2 protease cleavage site of the SARS-CoV-2 virus (FIGURE 1) (13). The S1 region of the Spike protein is responsible for binding to the host cell ACE2 receptor, where the S2 region is responsible for fusion of the viral RNA and cellular membranes. Polybasic furin sites in hemagglutinin (HA) proteins are often found in highly virulent avian and human influenza viruses (10). The insertion of the furin site may augment the ability of this new SARS-CoV-2 to attach and invade human cells expressing ACE2 and CD147 receptors (12, 86).
FIGURE 1.
Plasmin(ogen) increases the pathogeneticity of COVID-19. Plasmin cleaves the S protein of SARS-CoV-2 extracellularly, increasing its ability to bind with angiotensin converting enzyme 2 (ACE2) receptors of host cells, and probably facilitating virus entry and fusion. Plasmin proteolytically breaks down excess fibrin to elevate D-dimer and other fibrin degradation products in both bronchoalveolar lavage fluid and plasma, which decreases platelets and results in hemorrhage. Plasmin also cleaves epithelial sodium channel (ENaC) subunits, located at the apical membranes of epithelial cells in the airway, lung, and kidney. This increases the ability of Na+ ions to enter epithelial cells resulting in hypertension and dehydration of the fluid lining lung airways and alveolar cells.
A. Furin Proteolytically Cleaves Other Viral Proteins
The envelope proteins of numerous viruses, such as human immunodeficiency virus (HIV), human and avian influenza, herpes, Epstein-Barr, leukemia, dengue, Ebola, hepatitis B, measles, West Nile, Zika, respiratory syncytial virus (RSV), SARS, MERS, and Marburg virus, are cleaved by intracellular furin-like proteases. This increases the ability of the viruses to enter host cells (13, 34, 56). Proteolysis at a conventional “XXXR/S” motif in the S2 region of the Spike protein may facilitate the entry of respiratory infectious viruses (such as RSV and influenza) into airway and alveolar epithelial cells (94). Both SARS and MERS have evolved an unusual two-step furin activation for fusion, suggestive of a role during the process of emergence into the human population (55). Amidst the cluster of the furin-cleaved viruses, SARS S protein is cleaved by airway proteases (trypsin, plasmin, and TMPRSS11a), expressed in human bronchial epithelial cells, subsequently enhancing pseudovirus entry via binding host ACE2 receptors (37). The protease cleavage sites are R667 in the S1 fragment and R797 in the S2 fragment of SARS virus. These two cleavage sites are preserved in other coronaviruses, HIV, human and avian influenza virus, human CMV and RSV, yellow fever virus, and Zika virus (13).
B. Cleavage of Coronavirus by Plasmin and Other Host Proteases
Single-cell profiling of human lung tissues (the LGEA portal: https://research.cchmc.org/pbge/lunggens/mainportal.html) reveals that furin is predominately expressed in human alveolar type II (AT2) cells in the respiratory system, while plasminogen, kallikrein, and trypsin are expressed in both airway and alveolar type I and II epithelial cells. Plasminogen is also expressed in endothelial cells. Cytosolic furin is enriched in the Golgi apparatus. The possibility for non-furin proteases to cleave viral envelope proteins is supported by the evidence that in furin-defective LoVo cells, the cleavage-dependent process of HIV gp160 is as efficient as in normal cell lines (60). We have demonstrated that plasmin is capable of cleaving furin sites in the γ subunit of human epithelial sodium channels (ENaC) (99).
The S protein of coronaviruses may be cleaved by plasmin, trypsin, cathepsins, elastase, and TMPRSS family members; cleavage of S protein may mediate enhancement of virus entry into bronchial epithelial cells (37). Plasmin also cleaves the S proteins of SARS-CoV in vitro (37). In addition, HCoV-HKU1 S proteins are cleaved by kallikrein in the S1/S2 region and mediate the entry of HCoV-HKU1 to nonpermissive rhabdomyosarcoma cells (54). The clinical relevance of non-furin cleavage remains unknown due to the paucity of in vivo evidence for the role of plasmin cleavage of SARS-CoV. Also, it remains to be demonstrated that the envelope proteins of SARS-CoV-2 strain are cleaved by plasmin (56).
The cleavage of influenza virus by plasmin is well characterized (5, 23, 42, 57, 73, 83, 96). Proteolysis of influenza HA proteins enables fusion with the host endosome. Acidification of the endosome promotes viral membrane fusion and activates the M2 ion channel, which pumps protons (H1) into the interior of the viral core to initiate uncoating of the M1 protein. Nuclear replication occurs, and viral gene products are transported to the plasma membrane for assembly. The fibrinolytic zymogen plasminogen (activated by urokinase or tissue-like plasminogen activator to generate plasmin) has been shown to cleave the influenza HA proteins (40, 75, 83). The HA cleavage site of A/WSN/1933 H1N1 influenza virus governs virus spread in a plasmin-dependent manner (75). Mini-plasmin, a plasmin fragment, is distributed predominantly in the epithelial cells of the bronchioles and potentiates the replication of both plasmin-sensitive and plasmin-insensitive influenza A virus strains, suggesting a pivotal role of plasmin in the spread and pathogenicity of the influenza virus (57). Additionally, kallikreins cleave and activate HA of the influenza virus H1, H2, and H3 subtypes (27). Similar to coronavirus and influenza viruses, plasmin, trypsin, thrombin, and furin enhance RSV-induced cytopathology (17). Local fibrin and clot formation are implicated in host defense against influenza virus infections (5), thus plasminogen may affect lung injury and repair by interfering with these processes (1).
Protease cleavage may enhance or decrease the activities of various proteins. For example, prostasin increases the activity (60–80%) of ENaC, whereas TMPRSS2 markedly decreases ENaC function and protein levels (15). In neural tissues, brain-derived neurotrophic factor precursor (proBDNF) is cleaved either intracellularly by furin-like proteases or extracellularly by plasmin or matrix metalloproteinases. However, plasmin, but not related proteases, cleaves proBDNF furin sites extracellularly (4). The inhibitory effects of TMPRSS2 could be corrected by serine protease inhibitors, such as camostat mesylate that has been approved for clinical use in Japan (98). The beneficial effects of camostat mesylate, an antiprotease, may be partially due to the inhibition of plasmin (98).
C. Proteolytical Cleavage and Pathogenicity
Extracellular cleavage of virus envelope fusion glycoproteins by host cellular proteases is a prerequisite for the infectivity of respiratory viruses. The presence of a polybasic cleavage site that can be cleaved by furin-like proteases is a signature of several highly pathogenic avian influenza viruses (82). Similarly, the S protein of SARS-CoV-2 harbors a furin cleavage site at the S1/S2 boundary. The almost ubiquitous and diverse expression of furin-like proteases could lead to increasing SARS-CoV-2 cell and tissue tropism and transmissibility, and enhance its pathogenicity (84).
Four viral proteins are essential for the pathogenesis of COVID-19. The S proteins bind to ACE2 receptors after being cleaved by furin-like proteases. The RNA-dependent RNA polymerase (RdRp) is responsible for replicating SARS-CoV-2 RNA genome. 3C-like and papain-like proteases cleave two polyproteins that are important for the packing new virions. Whether plasmin and other host proteases cleave additional viral proteins is not known.
IV. ELEVATED PLASMIN(OGEN) LEVELS IN COMORBID DISEASES OF COVID-19
A. Plasmin(ogen) in Hypertensive Patients
Plasmin is generated from the cleavage of plasminogen by either urokinase (uPA) or tissue-like plasminogen activator (tPA). In general, uPA is responsible for the plasmin in body fluid (BAL, urine, tear, pleural effusion, etc.), while circulating tPA proteolytically cleaves plasminogen in the plasma. The most frequent comorbidity observed in COVID-19 patients is hypertension followed by diabetes, chronic cardiovascular conditions, cerebrovascular diseases, COPD, and chronic kidney illnesses (80, 101). The plasminogen system is a druggable target in renal hypertension (76). Plasmin, a potent protease, cleaves up to 16 sites, including the cleavage sites for trypsin, chymotrypsin, prostasin, and elastases, of the human γ ENaC subunit (99). Elevated renal plasmin results in hypertension by cleaving ENaC in the collecting tubule, which increases salt retention, causing expanded circulating volume. Urinary excretion of plasmin(ogen) and urokinase directly correlates with urine albumin in hypertensive subjects (2). On the other hand, amiloride, an inhibitor of ENaC, lowers blood pressure and urine plasminogen excretion (61). ENaC proteins are located in the apical membranes of tight epithelia, and they are the major pathways for the entry of Na+. As such, they play an important role in maintaining the proper depth of airway and alveolar lining fluids, the reabsorption of edema fluid in injured lungs, and the regulation of salt retention in the collecting tubules (6, 14, 18, 28, 38, 39, 50, 64). Proteolysis is an important regulatory mechanism of ENaC function (36, 63, 65, 67, 70, 99). Decreased ENaC function will result in increased fluid in body cavities (e.g., lung edema in the airspaces and increased blood volume), while increased function will cause dehydration of luminal fluid, as it occurs in cystic fibrosis (CF) and most likely dry eye syndrome.
B. Plasmin(ogen) in Cardiovascular Diseases
Significantly higher levels of urinary plasminogen and plasmin are reported in rats (71) and patients with chronic heart failure (76, 100). In addition, plasmin activity in patients with coronary artery disease is 1.7-fold greater compared with healthy subjects (16).
C. Plasmin(ogen) in Diabetes
Both types I and II diabetes are associated with higher plasmin(ogen) levels in plasma. A 25-year prospective study of type I diabetes documented an association with increased urinary plasmin(ogen), particularly in hypertensive subjects (69). Concentrations of plasmin(ogen) in urine are correlated with the development of preeclampsia late in pregnancy (58). Aberrant plasmin in preurine may inappropriately activate ENaC in patients with type II diabetes and microalbuminuria (7). Individuals with high plasma furin concentration have a pronounced dysmetabolic phenotype and elevated risk of diabetes mellitus and premature mortality (19).
D. Plasmin(ogen) in Other Comorbid Diseases
Higher levels of plasmin(ogen) are detected in the urine of various cancer patients as compared with healthy individuals (9). Elevated urine plasmin(ogen) levels, accompanied by increased exosomal α ENaC fragments, have been detected in pregnant women (59), a population susceptible to influenza (22). Endogenous channel-activating proteases, as well as proteases released by inflammatory cells (trypsin, elastase), activate ENaC either by cleaving critical amino acids in α and γ ENaC subunits, or by activating signaling pathways (66, 72). Aprotinin, a potent and reversible Kunitz-type inhibitor of several serine proteases, including trypsin, plasmin, and kallikreins, has been reported to inhibit sodium transport among a variety of epithelial cells (66). Other Kunitz-type serine protease inhibitors, such as hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 (placental bikunin), have also been demonstrated to inhibit prostasin and ENaC activity (77). Finally, a1-antitrypsin, an acute-phase glycoprotein and a member of the serine protease inhibitor (SERPIN) superfamily, inhibits ENaC in vitro and in vivo by decreasing protease activity (41). Of note, SARS S protein inhibits ENaC via the protein kinase C signaling pathway (35). It is worth noting that high plasmin levels may contribute to the development of comorbid bacteremia and sepsis (26, 74). Interestingly, azithromycin, a common antibiotic for suppressing infection in the CF airways when combined with hydroxychloroquine, turns COVID-19 to SARS-CoV-2 negative in 5 days (21).
V. PLASMIN(OGEN) IN ARDS
A. Plasmin(ogen) Is Increased in ARDS
ARDS is a life-threatening disorder associated with respiratory and systemic infections, trauma, burns, inhalation of toxic gases, and aspiration of gastric contents injury (52). In addition to lung injury, patients with ARDS may develop MOF with a hallmark of excess D-dimer and other FDPs presenting in both BAL and blood biopsies. Soluble D-dimer and D-monomer are predominately produced from the proteolytic cleavage of cross-linked fibrin and fibrinogen/non-cross-linked fibrin, respectively, by plasmin (fibrinolysis). Plasmin activity in BAL is detected in healthy subjects (71). Both D-dimer and D-monomer levels are significantly increased up to 17-fold in undiluted edema fluid in patients with ARDS (68). Significant increase in both plasminogen and cleaved plasmin protein in the BAL of ARDS patients (32) and an animal model of DAD (33) have been reported. Augmented plasmin activity contributes to elevated D-dimer in the BAL of infected lungs in a time-dependent manner during the development of ARDS (20, 87). This is further validated by the observation that plasmin-mediated fibrinolytic activity could be inhibited by 50% with α2-antiplasmin antibody (32). Kallikrein and neutrophil elastase may contribute to the residual proteolytic activity in BAL of ARDS (53). Also, radiation-induced lung injury in mesothelioma patients is accompanied by a significant elevation in BAL plasminogen and plasmin-associated fibrinolytic activity (49).
B. Fibrinolysis in COVID-19
In comparison with patients with mild COVID-19 (such as those who did not require ICU stays, did not develop ARDS or pneumonia, and who survived), patients with severe COVID-19 have higher comorbidities, including 56% for hypertension, 21% for heart diseases, 18% for diabetes, 12% for cerebrovascular diseases, and 7% for cancer (TABLE 1) (79, 97). Some patients have more than one, even up to five preexisting conditions. Multivariate regression further links hypertension with increased incidence and fatality (85, 89, 101). Hyperfibrinolysis, reflected by elevated serum D-dimer levels, was present in 97% of COVID-19 patients at admission and increased further in all patients before death (TABLE 2) (97). FDPs were significantly increased as well (79). This is accompanied by a prolonged prothrombin time particularly in non-survivors (31, 79, 97, 101). Platelet counts were decreased significantly in severe and dead patients (79, 97, 101). 71.4% of non-survivors meet the criteria of the International Society on Thrombosis and Hemostasis (ISTH) for DIC, suggesting the coexistence of coagulation activation and hyperfibrinolysis in patients with severe COVID-19 infection (78, 85). In contrast, D-dimer levels decreased to control levels in survivors or non-ARDS patients.
Table 1.
Comorbidities of COVID-19 patients
| Epidemiology (death%) (n = 72,314) | Severe/non-severe Pneumonia (n = 38/72) | Severe/non-severe (n = 173/926) | ICU/non-ICU (n = 13/28) | ARDS/non-ARDS (n = 53/56) | Non-survivor/survivor (n = 54/137) | Non-survivor/survivor (n = 32/20) | Non-survivor (n = 82) | |
|---|---|---|---|---|---|---|---|---|
| Hypertension | 12.8/6 | 39.47/29.17c | 23.7/13.4 | 15/14 | 39.6/28.6 | 48/23%c | 56.1 | |
| Diabetes | 5.3/7.3 | 21.05/9.72 | 16.2/5.7 | 8/25 | 20.8/1.8b | 31/14b | 22/10 | 18.3 |
| Coronary heart diseases | 4.2/10.5 | 5.8/1.8 | 23/11 | 5.7/7.1 | 24/1d | 9/10 | 20.7 | |
| Cerebrovascular diseases | 7.89/5.56 | 2.3/1.2 | 11.3/0b | 22/0 | 12.2 | |||
| COPD | 2.4/6.3 | 10.53/2.78a | 3.5/0.6 | 8/0 | 3.8/3.6 | 7/1a | 6/10 | 14.6 |
| Kidney diseases | 1.7/0.5 | 15.1/3.6a | 4/0a | 4.9 | ||||
| Liver diseases | 0.6/2.4 | 0/1 | — | 2.4 | ||||
| Cancer | 0.5/5.6 | 1.7/0.8 | 0/1 | 0/1 | 3/5 | 7.3 | ||
| Secondary infection | 0.6/2.4 | 6.1 | ||||||
| Immunodeficiency | 0/0.2 | 17.1 | ||||||
| Others | 20/8a | 3.7 (surgery) | ||||||
| Total | 26/− | 38.7/21 | 38/29 | 67/40c | 76.8 | |||
| Reference no. | 80 | 85 | 24 | 31 | 45 | 101 | 95 | 97 |
ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease; ICU, intensive care unit.
P < 0.05,
P < 0.01,
P < 0.001,
P < 0.0001.
Table 2.
Coagulation and fibrinolysis in patients with COVID-19
| Severe/non-severe (n = 173/926) | ARDS/non-ARDS (n = 53/56) | ICU/non-ICU (n = 13/28) | Severe/non-severe Pneumonia (n = 38/72) | Non-survivor/survivor (n = 54/137) | Non-survivor/survivor (n = 21/162) | Non-survivor/survivor (n = 32/20) | Non-survivor (n = 82) | ||
|---|---|---|---|---|---|---|---|---|---|
| D-dimer, >1 mg/L | 59.6/43.2 (≥0.5 mg/L) | 940/370c | 2.4/0.5b | 1.11/0.37c | 5.2/0.6d | 2.12/0.61c, 100/ | 97.5–100 (>0.55 mg/L) | ||
| FDP, mg/L | 7.6/4.0c | ||||||||
| Fibrinogen, g/L | 3.4/2.9c | 5.16/4.51, 28.6 (<1 g/L) | — | ||||||
| Platelets, <100 × 109/L | 57.7/31.6 (<150,000/mm3) | 8/4 | 144.5/179.5c (109/L) | 20%/1%d | 57.1/ | 191/164 | 63.2 | ||
| Prothrombin time, ≥16 s | 12.2/10.7a | 13/3a | 15.5/13.6c | 100 (>12.1) | |||||
| Antithrombin activity | 84/91 | 12.9/10.9 | |||||||
| APTT, s | 26.2/27.7 | 44.8/41.2 | |||||||
| ISTH DIC criteria | 71.4/- | ||||||||
| Reference no. | 24 | 45 | 31 | 85 | 101 | 79 | 95 | 97 |
APTT, activated partial thromboplastin time; ARDS, acute respiratory distress syndrome; DIC, disseminated intravascular coagulation; FDP, fibrin degradation products; ICU, intensive care unit; ISTH, International Society on Thrombosis and Hemostasis.
P < 0.05,
P < 0.01,
P < 0.001,
P < 0.0001.
The mortality rate of patients with COVID-19 who did not develop ARDS is 9 versus 49% for those who did develop ARDS (45). Of note, ARDS/respiratory failure remains the leading cause of death (70%), followed by sepsis/MOF (28%), heart failure (15%), hemorrhage (6%), and renal failure (4%) (TABLE 3). Coagulation/hemorrhage ranks among the top three leading causes of death (97). Furthermore, multivariate regression analysis identifies D-dimer and age as independent risk factors for mortality (TABLE 4) (85, 89, 101). These findings suggest that the normalization of hyperactive fibrinolysis may be a therapeutic target.
Table 3.
Outcomes or complications (%) in patients with COVID-19
| Severe/non-severe (n = 173/926) | ICU/non-ICU (n = 13/28) | Non-survivor/survivor (n = 54/137) | Non-survivor/survivor (n = 32/20) | Injured Organs (n = 82) | Death Cause (n = 82) | |
|---|---|---|---|---|---|---|
| Sepsis/MOF | 100/42d | 3/0 | 28.0e | |||
| Respiratory failure | 98/36d | 100 | ||||
| ARDS | 16.5/1.1 | 85/4d | 93/7d | 81/45 | 69.5e | |
| Septic shock | 6.4/0.1 | 23/0a | 70/0d | |||
| Acute cardiac injury | 31/4a | 59/1d | 28/15 | 89.0 | ||
| Heart failure | 52/12d | 14.6e | ||||
| Coagulopathy/hemorrhage | 50/7d | 6.1e | 80.5 | |||
| Acute kidney injury | 2.9/0.1 | 23/0a | 50/1d | 37.5/15 | 31.7 | |
| Secondary infection | 31/0a | 50/1d | 9/20 | |||
| Hypoproteinemia | 37/1d | |||||
| Acidosis | 30/1d | 2.4e | ||||
| Renal failure | 0.6/0 | 3.7e | ||||
| Liver failure | 1.2e | 78.0 | ||||
| GI failure | 2.4e | 6.1 | ||||
| Reference no. | 24 | 31 | 101 | 77 | 97 | 97 |
ARDS, acute respiratory distress syndrome; GI, gastrointestinal; ICU, intensive care unit; MOF, multiple organ failure.
P < 0.05,
P < 0.01,
P < 0.001,
P < 0.0001.
Percent of contribution to death.
Table 4.
Risk factors of COVID-19 associated with mortality computed with multivariate logistic regression
| Non-survivor/survivor (OR, n = 54/137) | Severe/non-severe Pneumonia (OR, n = 38/72) | ARDS (HR, n = 201) |
|
|---|---|---|---|
| Age | 1.10 (1.03, 1.17)b | 25.314 (1.628, 92.664)c >60 yr | 6.17 (3.26, 11.67)c |
| Lymphocyte | 0.19 (0.01, 1.62) | 0.322 (0.137, 0.756)b | 0.51 (0.22, 1.17) |
| D-dimer | 18.42 (2.64, 128.55)b | 17.054 (2.547, 114.171)b | 1.02 (1.01, 1.04)b |
| Reference no. | 101 | 85 | 89 |
ARDS, acute respiratory distress syndrome; HR, hazard ratio; OR, odd ratio.
P < 0.05,
P < 0.01,
P < 0.001.
C. Uncoordinated Coexistence of Hypercoagulation and Hyperproteolysis
The specific plasmin inhibitor α2-antiplasmin is elevated by approximately one order of magnitude in patients with ARDS, while fibrinolytic activity is reduced approximately by half, and D-dimer is elevated 50-fold in BAL (25). The nonproportional change between the expression and activity level of plasmin(ogen) and anti-plasmin indicates the stoichiometry of the plasmin-antiplasmin complexes may not be in a ratio of 1:1. Increased levels of α2-antiplasmin and other antiproteases may not completely shield the proteolytic triad of plasmin in the complexes, suggesting that either their efficacy is inadequate or that plasmin is still able to cut fibrin to produce D-dimers and FDPs in ARDS patients. The soluble complexes of the plasmin-antiplasmin in BAL may facilitate physical interactions with the vast deposition of fibrin at the luminal surface of alveoli. Based on the pathology and laboratory results, dynamic hypercoagulation occurs as evidenced by microthrombi throughout the blood vessels of multiple organs, accompanied with extremely reduced platelets in COVID-19 patients (TABLE 2). On the other hand, hemorrhage and markedly elevated degraded fibrin products result from plasmin-associated hyperproteolysis. Whether patchy hemorrhage coexists with areas infected by SARS-CoV-2 is not known. Administration of anti-proteases may prove beneficial.
VI. CLINICAL RELEVANCE AND PERSPECTIVE
The cleavage of the new furin sites in the S protein of SARS-CoV-2 virus by plasmin and other proteases may enhance its infectivity by expediting entry, fusion, duplication, and release in respiratory cells. Elevated plasmin(ogen) levels are a common feature in COVID-19 patients with underlying medical conditions. The elevated plasmin(ogen) could be an independent factor for risk stratification of patients with COVID-19. Measurements of plasmin(ogen) levels and its enzymatic activity may be important biomarkers of disease severity in addition to resultant D-dimer. The administration of antiproteases to suppress plasmin activity in the respiratory system may prevent, or at least decrease, SARS-CoV-2 entry into respiratory cells and improve the clinical outcome of patients with COVID-19. As demonstrated in vitro, a serine protease inhibitor for TMPRSS2 blocks SARS-CoV-2 S protein-driven entry into cells (30). Clinical trials conducted in China are testing various protease inhibitors (29). Currently there are no proper animal models of COVID-19 with underlying medical conditions to test new therapeutic agents. Healthy mice and monkeys infected with SARS-CoV-2 develop either mild lung injury or show no symptoms of disease (8). It remains to be seen whether mice and monkeys with preexisting comorbid conditions and higher plasmin levels develop COVID-19 when infected with SARS-CoV-2. Targeting hyperfibrinolysis with a broad spectrum or specific anti-plasmin compounds may prove to be a promising strategy for improving the clinical outcome of patients with comorbid conditions.
GRANTS
This study was supported by National Institutes of Health Grants HL87017 (to H.-L. Ji), HL095435 and HL134828 (to M. A. Matthay), and 5U01ES026458 and 5U01ES027697 (to S. Matalon).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
We are grateful to Dr. Lee Ann Riesenberg (Univ. of Alabama at Birmingham) for her professional editing of the manuscript.
Address for reprint requests and other correspondence: H.-L. (James) Ji, Dept. of Cellular and Molecular Biology, Univ. of Texas Health Science Centre at Tyler, Tyler, TX 75708 (e-mail: James.ji@uthct.edu).
Footnotes
These numbers are increasing daily now.
REFERENCES
- 1.Ali G, Zhao R, Zhang M, Jain KG, Chang J, Komatsu S, Fang X, Zhou B, Liang J, Jiang D, Ikebe M, Matthay MA, Ji H-L. Fibrinolytic niche is requested for alveolar type 2 cell-mediated alveologenesis and injury repair. bioRxiv. doi: 10.1101/2020.03.24.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen H, Friis UG, Hansen PB, Svenningsen P, Henriksen JE, Jensen BL. Diabetic nephropathy is associated with increased urine excretion of proteases plasmin, prostasin and urokinase and activation of amiloride-sensitive current in collecting duct cells. Nephrol Dial Transplant 30: 781–789, 2015. doi: 10.1093/ndt/gfu402. [DOI] [PubMed] [Google Scholar]
- 3.Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M, Lee M. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington state. JAMA. In press. doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benicky J, Sanda M, Brnakova Kennedy Z, Goldman R. N-Glycosylation is required for secretion of the precursor to brain-derived neurotrophic factor (proBDNF) carrying sulfated LacdiNAc structures. J Biol Chem 294: 16816–16830, 2019. doi: 10.1074/jbc.RA119.009989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berri F, Rimmelzwaan GF, Hanss M, Albina E, Foucault-Grunenwald ML, Lê VB, Vogelzang-van Trierum SE, Gil P, Camerer E, Martinez D, Lina B, Lijnen R, Carmeliet P, Riteau B. Plasminogen controls inflammation and pathogenesis of influenza virus infections via fibrinolysis. PLoS Pathog 9: e1003229, 2013. doi: 10.1371/journal.ppat.1003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boscardin E, Alijevic O, Hummler E, Frateschi S, Kellenberger S. The function and regulation of acid-sensing ion channels (ASICs) and the epithelial Na+ channel (ENaC): IUPHAR Review 19. Br J Pharmacol 173: 2671–2701, 2016. doi: 10.1111/bph.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhl KB, Oxlund CS, Friis UG, Svenningsen P, Bistrup C, Jacobsen IA, Jensen BL. Plasmin in urine from patients with type 2 diabetes and treatment-resistant hypertension activates ENaC in vitro. J Hypertens 32: 1672–1677, 2014. doi: 10.1097/HJH.0000000000000216. [DOI] [PubMed] [Google Scholar]
- 8.Callaway E. Labs rush to study coronavirus in transgenic animals–some are in short supply. Nature 579: 183, 2020. doi: 10.1038/d41586-020-00698-x. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Veitonmaki N, Keough K, Cheng H, Lee LS, Zurakowski D. Elevated levels of urine angiostatin and plasminogen/plasmin in cancer patients. Int J Mol Med 5: 547–551, 2000. doi: 10.3892/ijmm.5.5.547. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95: 409–417, 1998. doi: 10.1016/S0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395: 507–513, 2020. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J, Xing J, Shang P, Qian A, Li Y, Shaw PX, Wang J, Duan S, Ding J, Fan C, Zhang Y, Yang Y, Yu X, Feng Q, Li B, Yao X, Zhang Z, Li L, Xue X, Zhu P. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis 191: 755–760, 2005. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176: 104742, 2020. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Y, Zhao R, Zhao X, Matthay MA, Nie HG, Ji HL. ENaCs as both effectors and regulators of miRNAs in lung epithelial development and regeneration. Cell Physiol Biochem 44: 1120–1132, 2017. doi: 10.1159/000485417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. Regulation of the epithelial sodium channel by serine proteases in human airways. J Biol Chem 277: 8338–8345, 2002. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- 16.Drinane MC, Sherman JA, Hall AE, Simons M, Mulligan-Kehoe MJ. Plasminogen and plasmin activity in patients with coronary artery disease. J Thromb Haemost 4: 1288–1295, 2006. doi: 10.1111/j.1538-7836.2006.01979.x. [DOI] [PubMed] [Google Scholar]
- 17.Dubovi EJ, Geratz JD, Tidwell RR. Enhancement of respiratory syncytial virus-induced cytopathology by trypsin, thrombin, and plasmin. Infect Immun 40: 351–358, 1983. doi: 10.1128/IAI.40.1.351-358.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaton DC, Helms MN, Koval M, Bao HF, Jain L. The contribution of epithelial sodium channels to alveolar function in health and disease. Annu Rev Physiol 71: 403–423, 2009. doi: 10.1146/annurev.physiol.010908.163250. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez C, Rysä J, Almgren P, Nilsson J, Engström G, Orho-Melander M, Ruskoaho H, Melander O. Plasma levels of the proprotein convertase furin and incidence of diabetes and mortality. J Intern Med 284: 377–387, 2018. doi: 10.1111/joim.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs-Buder T, de Moerloose P, Ricou B, Reber G, Vifian C, Nicod L, Romand JA, Suter PM. Time course of procoagulant activity and D dimer in bronchoalveolar fluid of patients at risk for or with acute respiratory distress syndrome. Am J Respir Crit Care Med 153: 163–167, 1996. doi: 10.1164/ajrccm.153.1.8542111. [DOI] [PubMed] [Google Scholar]
- 21.Gautret P, Lagier J, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Esteves V, Dupont HT, Honoré S, Colson P, Chabrière E, Scola BL, Rolain J, Brouqui P, Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. In press. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldkind SF, Sahin L, Gallauresi B. Enrolling pregnant women in research–lessons from the H1N1 influenza pandemic. N Engl J Med 362: 2241–2243, 2010. doi: 10.1056/NEJMp1003462. [DOI] [PubMed] [Google Scholar]
- 23.Goto H, Wells K, Takada A, Kawaoka Y. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J Virol 75: 9297–9301, 2001. doi: 10.1128/JVI.75.19.9297-9301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19 . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. In press. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Günther A, Mosavi P, Heinemann S, Ruppert C, Muth H, Markart P, Grimminger F, Walmrath D, Temmesfeld-Wollbrück B, Seeger W. Alveolar fibrin formation caused by enhanced procoagulant and depressed fibrinolytic capacities in severe pneumonia. Comparison with the acute respiratory distress syndrome. Am J Respir Crit Care Med 161: 454–462, 2000. doi: 10.1164/ajrccm.161.2.9712038. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Li J, Hagström E, Ny T. Beneficial and detrimental effects of plasmin(ogen) during infection and sepsis in mice. PLoS One 6: e24774, 2011. doi: 10.1371/journal.pone.0024774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton BS, Whittaker GR. Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J Biol Chem 288: 17399–17407, 2013. doi: 10.1074/jbc.M112.440362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanukoglu I, Hanukoglu A. Epithelial sodium channel (ENaC) family: phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene 579: 95–132, 2016. doi: 10.1016/j.gene.2015.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. In press. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 1–10, 2020. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497–506, 2020. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Idell S, James KK, Levin EG, Schwartz BS, Manchanda N, Maunder RJ, Martin TR, McLarty J, Fair DS. Local abnormalities in coagulation and fibrinolytic pathways predispose to alveolar fibrin deposition in the adult respiratory distress syndrome. J Clin Invest 84: 695–705, 1989. doi: 10.1172/JCI114217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Idell S, Peters J, James KK, Fair DS, Coalson JJ. Local abnormalities of coagulation and fibrinolytic pathways that promote alveolar fibrin deposition in the lungs of baboons with diffuse alveolar damage. J Clin Invest 84: 181–193, 1989. doi: 10.1172/JCI114139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izaguirre G. The proteolytic regulation of virus cell entry by Furin and other proprotein convertases. Viruses 11: 837, 2019. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji H-L, Song W, Gao Z, Su X-F, Nie H-G, Jiang Y, Peng J-B, He Y-X, Liao Y, Zhou Y-J, Tousson A, Matalon S. SARS-CoV proteins decrease levels and activity of human ENaC via activation of distinct PKC isoforms. Am J Physiol Lung Cell Mol Physiol 296: L372–L383, 2009. doi: 10.1152/ajplung.90437.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji HL, Zhao R, Komissarov AA, Chang Y, Liu Y, Matthay MA. Proteolytic regulation of epithelial sodium channels by urokinase plasminogen activator: cutting edge and cleavage sites. J Biol Chem 290: 5241–5255, 2015. doi: 10.1074/jbc.M114.623496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kam YW, Okumura Y, Kido H, Ng LF, Bruzzone R, Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One 4: e7870, 2009. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleyman TR, Kashlan OB, Hughey RP. Epithelial Na+ channel regulation by extracellular and intracellular factors. Annu Rev Physiol 80: 263–281, 2018. doi: 10.1146/annurev-physiol-021317-121143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kone BC. Epigenetics and the control of the collecting duct epithelial sodium channel. Semin Nephrol 33: 383–391, 2013. doi: 10.1016/j.semnephrol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazarowitz SG, Compans RW, Choppin PW. Proteolytic cleavage of the hemagglutinin polypeptide of influenza virus. Function of the uncleaved polypeptide HA. Virology 52: 199–212, 1973. doi: 10.1016/0042-6822(73)90409-1. [DOI] [PubMed] [Google Scholar]
- 41.Lazrak A, Nita I, Subramaniyam D, Wei S, Song W, Ji H-L, Janciauskiene S, Matalon S. Alpha(1)-antitrypsin inhibits epithelial Na+ transport in vitro and in vivo. Am J Respir Cell Mol Biol 41: 261–270, 2009. doi: 10.1165/rcmb.2008-0384OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeBouder F, Lina B, Rimmelzwaan GF, Riteau B. Plasminogen promotes influenza A virus replication through an annexin 2-dependent pathway in the absence of neuraminidase. J Gen Virol 91: 2753–2761, 2010. doi: 10.1099/vir.0.023804-0. [DOI] [PubMed] [Google Scholar]
- 43.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, Li C, Ai Q, Lu W, Liang H, Li S, He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 21: 335–337, 2020. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Wang R, Qu G, Wang Y, Liu P, Zhu Y, Fei G, Ren L, Zhou Y, Liu L. Anatomic examination of one dead COVID-19 patient. J Forensic Med 36: 19–21, 2020. [Google Scholar]
- 45.Liu Y, Sun W, Li J, Chen L, Wang Y, Zhang L, Yu L. Clinical features and progression of acute respiratory distress syndrome in coronavirus disease 2019. medRxiv. doi: 10.1101/2020.02.17.20024166. [DOI] [Google Scholar]
- 46.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, Peiris M, Poon LLM, Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. In press. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395: 565–574, 2020. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo W, Yu H, Gou J, Li X, Sun Y, Li J, Liu L. Clinical pathology of critical patient with novel coronavirus pneumonia (COVID-19). Preprints 2020: 2020020407, 2020. [Google Scholar]
- 49.Maasilta P, Salonen EM, Vaheri A, Kivisaari L, Holsti LR, Mattson K. Procollagen-III in serum, plasminogen activation and fibronectin in bronchoalveolar lavage fluid during and following irradiation of human lung. Int J Radiat Oncol Biol Phys 20: 973–980, 1991. doi: 10.1016/0360-3016(91)90193-8. [DOI] [PubMed] [Google Scholar]
- 50.Matalon S, Bartoszewski R, Collawn JF. Role of epithelial sodium channels in the regulation of lung fluid homeostasis. Am J Physiol Lung Cell Mol Physiol 309: L1229–L1238, 2015. doi: 10.1152/ajplung.00319.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matthay MA, Aldrich JM, Gotts JE. Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Respir Med S2213-2600(20)30127-2, 2020. doi: 10.1016/S2213-2600(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, Herridge M, Randolph AG, Calfee CS. Acute respiratory distress syndrome. Nat Rev Dis Primers 5: 18, 2019. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuire WW, Spragg RG, Cohen AB, Cochrane CG. Studies on the pathogenesis of the adult respiratory distress syndrome. J Clin Invest 69: 543–553, 1982. doi: 10.1172/JCI110480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milewska A, Falkowski K, Kalinska M, Bielecka E, Naskalska A, Mak P, Lesner A, Ochman M, Urlik M, Potempa J, Kantyka T, Pyrc K. Kallikrein 13: a new player in coronaviral infections. bioRxiv. doi: 10.1101/2020.03.01.971499. [DOI] [Google Scholar]
- 55.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA 111: 15214–15219, 2014. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202: 120–134, 2015. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, Parry MA, Kido H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem 268: 2847–2855, 2001. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen LH, Jensen BL, Fuglsang J, Andersen LLT, Jensen DM, Jørgensen JS, Kitlen G, Ovesen P. Urine albumin is a superior predictor of preeclampsia compared to urine plasminogen in type I diabetes patients. J Am Soc Hypertens 12: 97–107, 2018. doi: 10.1016/j.jash.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Nielsen MR, Frederiksen-Møller B, Zachar R, Jørgensen JS, Hansen MR, Ydegaard R, Svenningsen P, Buhl K, Jensen BL. Urine exosomes from healthy and hypertensive pregnancies display elevated level of α-subunit and cleaved α- and γ-subunits of the epithelial sodium channel-ENaC. Pflugers Arch 469: 1107–1119, 2017. doi: 10.1007/s00424-017-1977-z. [DOI] [PubMed] [Google Scholar]
- 60.Ohnishi Y, Shioda T, Nakayama K, Iwata S, Gotoh B, Hamaguchi M, Nagai Y. A furin-defective cell line is able to process correctly the gp160 of human immunodeficiency virus type 1. J Virol 68: 4075–4079, 1994. doi: 10.1128/JVI.68.6.4075-4079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oxlund CS, Buhl KB, Jacobsen IA, Hansen MR, Gram J, Henriksen JE, Schousboe K, Tarnow L, Jensen BL. Amiloride lowers blood pressure and attenuates urine plasminogen activation in patients with treatment-resistant hypertension. [Correction in J Am Soc Hypertens 9: 76, 2015.] J Am Soc Hypertens 8: 872–881, 2014. doi: 10.1016/j.jash.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 62.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis S1473-3099(20)30113-4, 2020. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Passero CJ, Hughey RP, Kleyman TR. New role for plasmin in sodium homeostasis. Curr Opin Nephrol Hypertens 19: 13–19, 2010. doi: 10.1097/MNH.0b013e3283330fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavlov TS, Staruschenko A. Involvement of ENaC in the development of salt-sensitive hypertension. Am J Physiol Renal Physiol 313: F135–F140, 2017. doi: 10.1152/ajprenal.00427.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Planès C, Caughey GH. Regulation of the epithelial Na+ channel by peptidases. Curr Top Dev Biol 78: 23–46, 2007. doi: 10.1016/S0070-2153(06)78002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Planès C, Leyvraz C, Uchida T, Angelova MA, Vuagniaux G, Hummler E, Matthay M, Clerici C, Rossier B. In vitro and in vivo regulation of transepithelial lung alveolar sodium transport by serine proteases. Am J Physiol Lung Cell Mol Physiol 288: L1099–L1109, 2005. doi: 10.1152/ajplung.00332.2004. [DOI] [PubMed] [Google Scholar]
- 67.Planès C, Randrianarison NH, Charles RP, Frateschi S, Cluzeaud F, Vuagniaux G, Soler P, Clerici C, Rossier BC, Hummler E. ENaC-mediated alveolar fluid clearance and lung fluid balance depend on the channel-activating protease 1. EMBO Mol Med 2: 26–37, 2010. doi: 10.1002/emmm.200900050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prabhakaran P, Ware LB, White KE, Cross MT, Matthay MA, Olman MA. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am J Physiol Lung Cell Mol Physiol 285: L20–L28, 2003. doi: 10.1152/ajplung.00312.2002. [DOI] [PubMed] [Google Scholar]
- 69.Ray EC, Miller RG, Demko JE, Costacou T, Kinlough CL, Demko CL, Unruh ML, Orchard TJ, Kleyman TR. Urinary plasmin(ogen) as a prognostic factor for hypertension. Kidney Int Rep 3: 1434–1442, 2018. doi: 10.1016/j.ekir.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossier BC, Stutts MJ. Activation of the epithelial sodium channel (ENaC) by serine proteases. Annu Rev Physiol 71: 361–379, 2009. doi: 10.1146/annurev.physiol.010908.163108. [DOI] [PubMed] [Google Scholar]
- 71.Sepper R, Konttinen YT, Buø L, Eklund KK, Lauhio A, Sorsa T, Tschesche H, Aasen AO, Sillastu H. Potentiative effects of neutral proteinases in an inflamed lung: relationship of neutrophil procollagenase (proMMP-8) to plasmin, cathepsin G and tryptase in bronchiectasis in vivo. Eur Respir J 10: 2788–2793, 1997. doi: 10.1183/09031936.97.10122788. [DOI] [PubMed] [Google Scholar]
- 72.Snyder PM. Minireview: regulation of epithelial Na+ channel trafficking. Endocrinology 146: 5079–5085, 2005. doi: 10.1210/en.2005-0894. [DOI] [PubMed] [Google Scholar]
- 73.Su H, Yang X, Wang S, Shi H, Liu X. Effect of annexin II-mediated conversion of plasmin from plasminogen on airborne transmission of H9N2 avian influenza virus. Vet Microbiol 223: 100–106, 2018. doi: 10.1016/j.vetmic.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjöbring U, Ginsburg D. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305: 1283–1286, 2004. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 75.Sun X, Tse LV, Ferguson AD, Whittaker GR. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J Virol 84: 8683–8690, 2010. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Svenningsen P, Hinrichs GR, Zachar R, Ydegaard R, Jensen BL. Physiology and pathophysiology of the plasminogen system in the kidney. Pflugers Arch 469: 1415–1423, 2017. doi: 10.1007/s00424-017-2014-y. [DOI] [PubMed] [Google Scholar]
- 77.Szabo R, Uzzun Sales K, Kosa P, Shylo NA, Godiksen S, Hansen KK, Friis S, Gutkind JS, Vogel LK, Hummler E, Camerer E, Bugge TH. Reduced prostasin (CAP1/PRSS8) activity eliminates HAI-1 and HAI-2 deficiency-associated developmental defects by preventing matriptase activation. PLoS Genet 8: e1002937, 2012. doi: 10.1371/journal.pgen.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost jth.14768, 2020. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Nat Sci Rev. In press. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) - China. Chin J Epidemiol. In press. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian S, Hu W, Niu L, Liu H, Xu H, Xiao S. Pulmonary pathology of early phase SARS-COV-2 pneumonia. Preprints 2020: 2020020220, 2020. doi: 10.20944/preprints202002.0220.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tse LV, Hamilton AM, Friling T, Whittaker GR. A novel activation mechanism of avian influenza virus H9N2 by furin. J Virol 88: 1673–1683, 2014. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tse LV, Marcano VC, Huang W, Pocwierz MS, Whittaker GR. Plasmin-mediated activation of pandemic H1N1 influenza virus hemagglutinin is independent of the viral neuraminidase. J Virol 87: 5161–5169, 2013. doi: 10.1128/JVI.00210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell S0092-8674(20)30262-2, 2020. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323: 1061, 2020. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, Gong L, Zhang Y, Cui H-Y, Geng J-J, Wang B, Sun X-X, Wang C-F, Yang X, Lin P, Deng Y-Q, Wei D, Yang X-M, Zhu Y-M, Zhang K, Zheng Z-H, Miao J-L, Guo T, Shi Y, Zhang J, Fu L, Wang Q-Y, Bian H, Zhu P, Chen Z-N. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woolley LK, Fell SA, Djordjevic SP, Eamens GJ, Jenkins C. Plasmin activity in the porcine airways is enhanced during experimental infection with Mycoplasma hyopneumoniae, is positively correlated with proinflammatory cytokine levels and is ameliorated by vaccination. Vet Microbiol 164: 60–66, 2013. doi: 10.1016/j.vetmic.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 88.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367: 1260–1263, 2020. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. In press. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature 579: 265–269, 2020. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72,314 cases from the Chinese Center for Disease Control and Prevention. JAMA. In press. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 92.Xia Y, Jin R, Zhao J, Li W, Shen H. Risk of COVID-19 for cancer patients. Lancet Oncol S1470-2045(20)30150-9, 2020. doi: 10.1016/S1470-2045(1020)30150-30159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. In press. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamada Y, Liu DX. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J Virol 83: 8744–8758, 2009. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. In press. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yao D, Kuwajima M, Kido H. Pathologic mechanisms of influenza encephalitis with an abnormal expression of inflammatory cytokines and accumulation of mini-plasmin. J Med Invest 50: 1–8, 2003. [PubMed] [Google Scholar]
- 97.Zhang B, Zhou X, Qiu Y, Feng F, Feng J, Jia Y, Zhu H, Hu K, Liu J, Liu Z, Wang S, Gong Y, Zhou C, Zhu T, Cheng Y, Liu Z, Deng H, Tao F, Ren Y, Cheng B, Gao L, Wu X, Yu L, Huang Z, Mao Z, Song Q, Zhu B, Wang J. Clinical characteristics of 82 death cases with COVID-19. medRxiv. doi: 10.1101/2020.02.26.20028191. [DOI] [Google Scholar]
- 98.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46: 586–590, 2020. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao R, Ali G, Nie HG, Chang Y, Bhattarai D, Su X, Zhao X, Matthay MA, Ji HL. Plasmin improves oedematous blood-gas barrier by cleaving epithelial sodium channels. Br J Pharmacol. In press. doi: 10.1111/bph.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng H, Liu X, Sharma NM, Li Y, Pliquett RU, Patel KP. Urinary proteolytic activation of renal epithelial Na+ channels in chronic heart failure. Hypertension 67: 197–205, 2016. doi: 10.1161/HYPERTENSIONAHA.115.05838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395: 1054–1062, 2020. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273, 2020. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733, 2020. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382: 1177–1179, 2020. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]