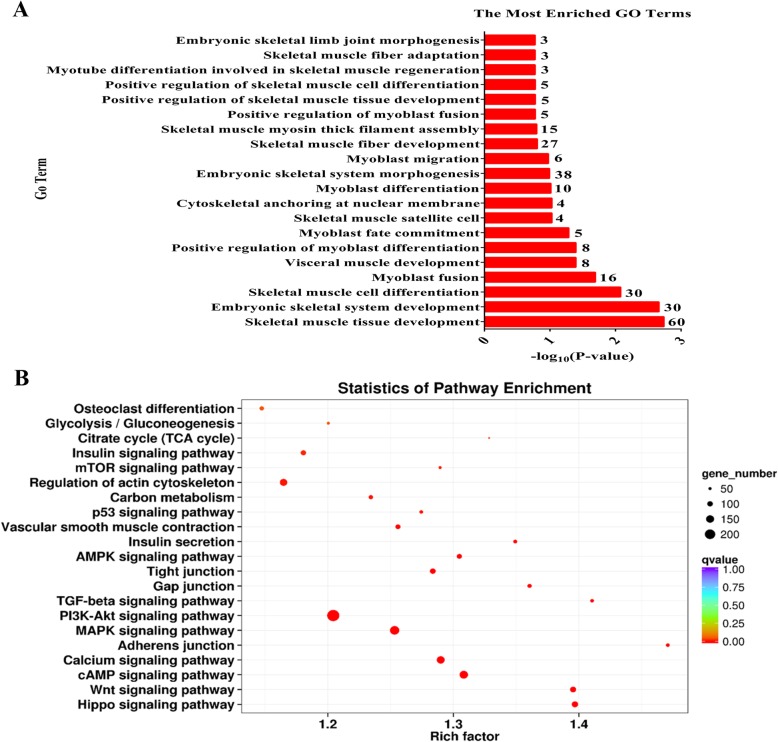
Fig. 8.
Enrichment analysis of CG-type differentially methylated genes. (a) Counts of DMGs enriched for the top 20 muscle development-related Gene Ontology (GO) terms. The abscissa represents the number of DMGs, and the ordinate shows the GO pathway terms. (b) The scatterplot of 21 muscle development-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The ordinate represents the enriched pathways, and the abscissa represents the Rich factor of the corresponding pathways; the size of the spots represents the number of genes related to DMRs enriched in each pathway, while the color of the spot represents the corrected P-value for each pathway. The Rich factors indicate the ratio of the number of DMGs mapped to a certain pathway to the total number of genes mapped to this pathway. Greater Rich factor means greater enrichment. DMRs: differentially methylated regions; DMGs: differentially methylated genes