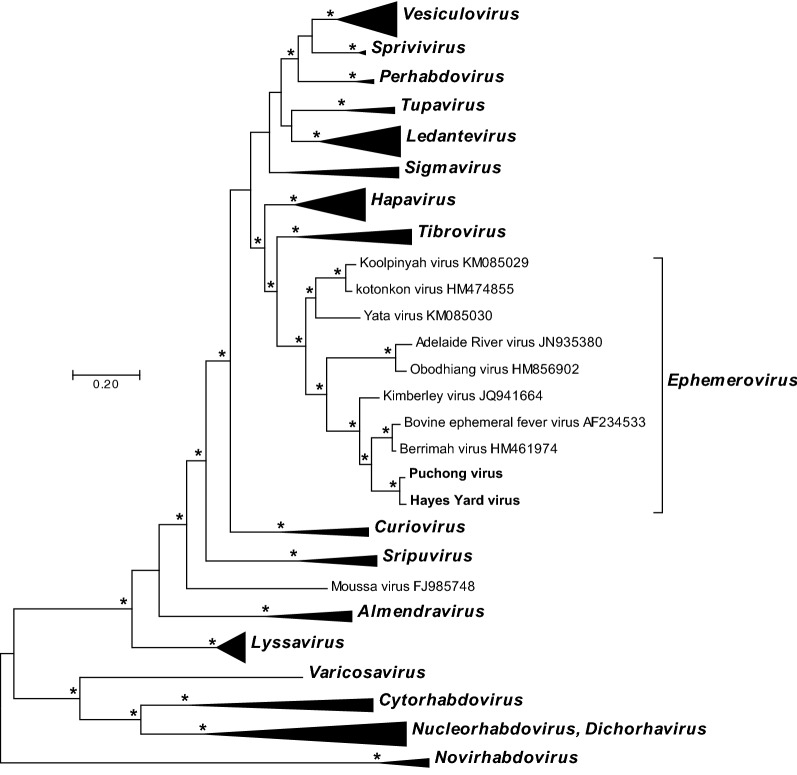
Figure 5.
Phylogenetic analysis of HYV and PUCV. A phylogenetic tree inferred from a MUSCLE alignment of complete L protein sequences of 126 rhabdoviruses currently assigned to species, as well as HYV and PUCV. Phylogenetically informative sites were selected from the alignment using Gblocks resulting in 565 positions in the final dataset. The tree was inferred in MEGA version 7.0.18 by using the Maximum Likelihood method. The tree with the highest log likelihood (−59 565.1689) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Nodes with bootstrap support values (1000 iterations) >70% are indicated (*). (Note that viruses currently assigned to the genus Nucleorhabdovirus and the genus Dichorhavirus are shown as a single cluster as the genera do not represent monophyletic groups).