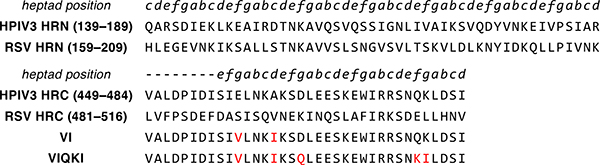Abstract
Human parainfluenza virus 3 (HPIV3) and respiratory syncytial virus (RSV) cause lower respiratory infection in infants and young children. There are no vaccines for these pathogens, and existing treatments have limited or questionable efficacy. Infection by HPIV3 or RSV requires fusion of the viral and cell membranes, a process mediated by a trimeric fusion glycoprotein (F) displayed on the viral envelope. Once triggered, the prefusion form of F undergoes a series of conformational changes that first extend the molecule to allow for insertion of the hydrophobic fusion peptide into the target cell membrane and then refold the trimeric assembly into an energetically stable postfusion state, a process that drives the merger of the viral and host cell membranes. Peptides derived from defined regions of HPIV3 F inhibit infection by HPIV3 by interfering with the structural transitions of the trimeric F assembly. Here we describe lipopeptides derived from the C-terminal heptad repeat (HRC) domain of HPIV3 F that potently inhibit infection by both HPIV3 and RSV. The lead peptide inhibits RSV infection as effectively as does a peptide corresponding to the RSV HRC domain itself. We show that the inhibitors bind to the N-terminal heptad repeat (HRN) domains of both HPIV3 and RSV F with high affinity. Co-crystal structures of inhibitors bound to the HRN domains of HPIV3 or RSV F reveal remarkably different modes of binding in the N-terminal segment of the inhibitor.
Introduction
Acute respiratory infection is the leading cause of mortality in young children. Most cases of childhood croup, bronchiolitis, and pneumonia, the most common manifestations of acute respiratory infections in infants, are caused by two viruses— human parainfluenza virus 3 (HPIV3) and respiratory syncytial virus (RSV).1,2 HPIV3 is a paramyxovirus that is responsible for 30–40% of all acute respiratory tract infections in infants and children.3 RSV is a pneumovirus that, as recently as 2015, infected approximately 33.1 million children under five, resulting in an estimated 3.2 million hospital admissions and nearly 60,000 in-hospital deaths.4 However, no antiviral agents are available to treat HPIV3 or RSV infections. Those at highest risk for severe infection can be treated prophylactically with a humanized monoclonal antibody against RSV, but this approach is costly and ineffective against existing infections.5 Given the similarity of symptoms caused by HPIV3 and RSV, antigen detection or PCR analysis is often required to identify the pathogen. Therefore, administration of an antiviral drug directed at one of these viruses would be delayed by the time required for diagnosis. An ideal drug would be effective against both HPIV3 and RSV, which would enable treatment at the onset of disease. Here we describe a single agent that blocks infectivity by both HPIV3 and RSV, which could provide a foundation for future therapeutic development efforts.
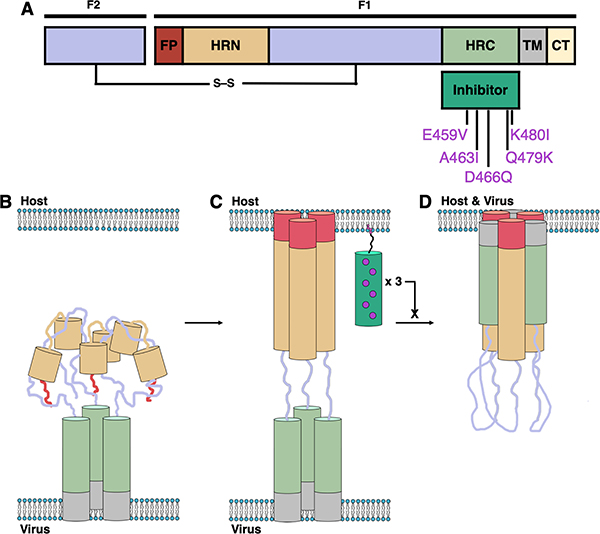
HPIV3 and RSV employ trimeric fusion (F) proteins to mediate merger of the viral envelope with the host cellular membrane, the first step of infection.6–8 F-induced membrane fusion is thought to proceed via a regulated sequence of structural transitions, as shown in Fig. 1. Prior to activation, the F protein trimer is anchored to the viral envelope by the transmembrane (TM) segments, each of which is adjacent to the C-terminal heptad repeat (HRC) domain. In this prefusion state, the N-terminal heptad repeat (HRN) domains and adjacent hydrophobic fusion peptide (FP) segments are folded into the head region of the trimeric assembly. For HPIV3, F protein activation is mediated by the receptor-engaged attachment protein HN; for RSV, the activation mechanism is unknown. Activation causes the HRN domains to extend, and the FP segments are inserted into the host cell membrane to form a transient “prehairpin” intermediate. An extensive structural change in the trimeric F assembly then brings the HRC and HRN domains together to form a compact and very stable six-helix bundle (6HB), which drives membrane fusion.
Figure 1.
Mechanism and inhibition of Class I fusion proteins. (A) Schematic of the protein with domains indicated: fusion peptide (FP, red); N-terminal heptad repeat (HRN, orange); C-terminal heptad repeat (HRC, green); transmembrane (TM, gray); cytoplasmic tail (CT, yellow). (B–D) Class I fusion mechanism depicting prefusion (B), prehairpin intermediate (C), and postfusion (D) conformations of F and the inhibition of fusion by peptide inhibitors (dark green).
Because viral entry is the essential first step of infection, the inhibition of F protein structural transitions is a potential strategy for the development of antiviral therapies.9 In several cases, a peptide based on a specific HRC domain has been shown to inhibit fusion mediated by the F protein from which the peptide was derived but not fusion mediated by F proteins of any other virus (e.g., enfuvirtide, an FDA-approved peptide drug derived from the HRC domain of the HIV fusion glycoprotein gp41, is effective against HIV but not effective against other viruses).10–14 Peptides derived from the HRC of HPIV3, however, exhibit activity against not only HPIV3 but also other paramyxoviruses including Nipah virus (NiV), parainfluenza virus 5 (PIV5) and measles virus (MeV).15–19 Antiviral potency requires that the peptide be conjugated to a hydrophobic moiety, such as cholesterol, which is thought to cause partitioning into membranes. These fusion-inhibitory peptides are hypothesized to act by binding to the exposed HRN domains of the F protein trimer in the extended prehairpin state.19–21 Despite the conserved fusion mechanism, the F proteins of different viruses possess distinct primary sequences; therefore, the surprising ability of HPIV3 HRC-derived peptides to disrupt the fusion machinery of multiple paramyxoviruses is potentially important from a therapeutic perspective. The potential benefit of HPIV3 HRC-derived peptides could be enhanced if the scope could be expanded to include potent inhibition of RSV, a member of the Pneumovirus family that often causes clinical presentation indistinguishable from that of HPIV3 in young children.
Here we report a cholesterol-conjugated peptide that inhibits infection of human cells by the major pediatric pathogens HPIV3 and RSV. The sequence is based on the HRC domain of HPIV3 F and contains five substitutions: E459V, A463I, D466Q, Q479K and K480I (residue numbering here and below based on the full-length HPIV3 F protein). This peptide is designated “VIQKI” below. X-ray co-crystal structures have been determined for the six-helix bundle assembly formed by VIQKI with a peptide derived from the HPIV3 HRN domain and the six-helix bundle assembly formed by VIQKI with a peptide derived from the RSV HRN domain. These structures reveal that the N-terminal segment of VIQKI adopts profoundly different conformations in these two assemblies. Alanine-scanning data support the conclusion that the crystal structures represent the binding modes responsible for inhibition of HPIV3 or RSV infectivity. Thus, our results offer unprecedented molecular-level insight on the mechanism by which a single peptide can adapt itself to distinct pathogen targets.
Results
Design and synthesis of sequence-optimized fusion inhibitory peptides
The fusion-inhibitory activity of HRC-derived peptides is hypothesized to result from inhibitor binding to the HRN domain of F in the prehairpin intermediate. We therefore sought an HRC-derived sequence that could engage the trimeric HRN assembly from both HPIV3 and RSV. We have previously reported that a 36-mer peptide corresponding to residues 449–484 of the HPIV3 HRC domain with two mutations, E459V and A463I (designated “VI”, Fig. 2), inhibits fusion by several paramyxoviruses, but not RSV.20 The VI peptide was used as a starting point for development of a fusion inhibitor active toward both HPIV3 and RSV. HRC-HRN association is primarily driven by hydrophobic interactions. Therefore, we made the change K480I to increase the local hydrophobicity of the HRC peptide at a site expected to be buried in the six-helix bundle formed with the HRN peptide. Other changes were intended to stabilize α-helical secondary structure, as required for six-helix bundle formation. Substitution D466Q was intended to remove a potentially repulsive i,i+3 Coulombic interaction with E469. Substitution Q479K was intended to encourage an i,i+3 salt bridge with D482. This last mutation increases the net charge of the peptide at neutral pH, which could counter the effects on peptide solubility of increased hydrophobicity at other sites.
Figure 2.
Aligned HRN- and HRC-derived peptide sequences. Peptide sequences of the aligned HRN and HRC domains of HPIV3 and RSV F and designed fusion inhibitory peptides VI and VIQKI are shown. Heptad repeat pattern for the sequences are indicated in italics.
Inhibitors form stable assemblies with HRN domains of HPIV3 and RSV F
When combined in solution, HRC- and HRN-derived peptides form a six-helix bundle assembly that is believed to mirror the helix-bundle within the postfusion state of F.19 We used circular dichroism (CD) spectroscopy to characterize assemblies that form between the native HRN and HRC domains of HPIV3 and RSV F, and to determine whether the VI and VIQKI peptides co-assemble with the HRN domains. At 50 μM in PBS, each of the four HRC peptides (HPIV3 HRC, RSV HRC, VI and VIQKI) exhibited minimal α-helical content. However, 1:1 mixtures of the native HRN/HRC partners (i.e., HPIV3 HRN+HPIV3 HRC and RSV HRN+RSV HRC) displayed intense minima at 208 and 222 nm (Fig. S33–S34), indicating a large increase in α-helical secondary structure relative to the individual peptides. This behavior indicates helix-bundle co-assembly of the HRC and HRN peptides. Heterotypic combinations of HRN and HRC peptides (i.e., HPIV3 HRN+RSV HRC or RSV HRN+HPIV3 HRC) exhibited double minima that are generally characteristic of helical assemblies. However, the minima observed for heterotypic pairings were significantly lower in intensity relative to the minima observed for native pairings, which suggests a smaller extent of α-helix formation for the cross-species pairs (Fig. S35–36, S41–42). The RSV HRN+HPIV3 HRC spectrum also showed a slight shift of the lower wavelength minimum to ~206 nm, and this minimum was significantly more intense relative to the 222 nm minimum (Fig. S42). These features suggest that some portions of the polypeptide backbone may be non-helical and perhaps unfolded in this assembly. Heating causes a relatively modest change in the intensity of the minimum at 222 nm (Fig. S45), with a transition presumed to reflect assembly disruption between 30 and 50 °C. The RSV HRN+VIQKI spectrum has a shape similar to that of RSV HRN+HPIV3 HRC (Fig. S42), but the minima are more intense (more negative) for the RSV HRN+VIQKI spectrum. This combination gives rise to a clearer thermal transition and a substantially higher temperature relative to the RSV HRN+HPIV3 HRC pairing (Fig. S50 vs. Fig. S45). These data indicate that the assembly formed by RSV HRN+VIQKI is more stable than the assembly formed by RSV HRN+HPIV3 HRC. Most importantly, these data suggest that VIQKI, an HPIV3-based peptide, can form six helix bundle assemblies with the HRN domains of the two distinct viral F proteins.
Variable-temperature CD studies were undertaken to compare stabilities among the co-assemblies. Each of the HRN+HRC pairs showed a highly cooperative thermal transition at 222 nm, which allowed determination of the apparent midpoint of thermal denaturation (Tm,app) values shown in Table 1. The thermal transitions are attributed to the disruption of the 6HB assembly with concomitant loss of α-helical secondary structure. Each Tm,app value, therefore, is a measure of the stability of the HRN+HRC co-assembly. To compare the designed inhibitors to the HPIV3 HRC from which they were derived, we calculated ΔTm,app values. In this case, ΔTm,app* is the difference between Tm,app for the inhibitor peptide in question co-assembled with the HPIV3 HRN peptide and the Tm,app for the native HPIV3 HRC peptide co-assembled with the HPIV3 HRN peptide. ΔTm,app† is the difference between Tm,app for the inhibitor peptide in question co-assembled with the RSV HRN peptide and the Tm,app for the native HPIV3 HRC peptide co-assembled with the RSV HRN peptide. Positive values indicate that the co-assembly of a particular inhibitor with either HPIV3 or RSV HRN is more stable than that of HPIV3 HRC. Negative values indicate the co-assembly is less stable than that of HPIV3 HRC.
Table 1.
Apparent melting temperatures (Tm,app) of coassemblies formed between HRC + HRN peptide pairs
| HPIV3 HRN | RSV HRN | |||
|---|---|---|---|---|
| Inhibitor | Tm,app (°C) | ΔTm,app* | Tm,app (°C) | ΔTm,app† |
| HPIV3 HRC | 86.8 | - | 42.2 | - |
| RSV HRC | 48.2 | −38.6 | 82.4 | +40.2 |
| VI | 88.1 | +1.3 | 56.9 | +14.7 |
| VIQKI | 88.6 | +1.8 | 64.3 | +22.1 |
ΔTm,app* = Tm,app (HPIV3 HRN+Inhibitor) − Tm,app (HPIV3 HRN+HPIV3 HRC)
ΔTm,app† = Tm,app (RSV HRN+Inhibitor) − Tm,app (RSV HRN+RSV HRC)
The native HPIV3 and RSV HRC peptides formed very stable assemblies with the natural HRN peptide partner (Tm,app = 86.8 and 82.4 °C, respectively). In contrast, the unnatural pairings, HPIV3 HRN+RSV HRC and RSV HRN+HPIV3 HRC, formed much less stable assemblies (Tm,app = 48.2 and 42.2 °C, respectively). The designed peptides VI and VIQKI formed assemblies with HPIV3 HRN that were comparable in stability to the HPIV3 HRC+HPIV3 HRN assembly. In contrast, the assemblies formed by VI and VIQKI with RSV HRN were substantially more stable (Tm,app = 56.9 and 64.3 °C, respectively) than the HPIV3 HRC+RSV HRN assembly. Thus, the substitutions that generate VI and VIQKI from HPIV3 HRC maintain co-assembly propensity with HPIV3 HRN while enhancing co-assembly propensity with RSV HRN. The behavior of VIQKI is particularly promising and suggests that this designed peptide could function as a dual inhibitor of HPIV3 and RSV infection, a hypothesis that is tested in the experiments described below.
Inhibition of fusion mediated by HPIV3 and RSV F
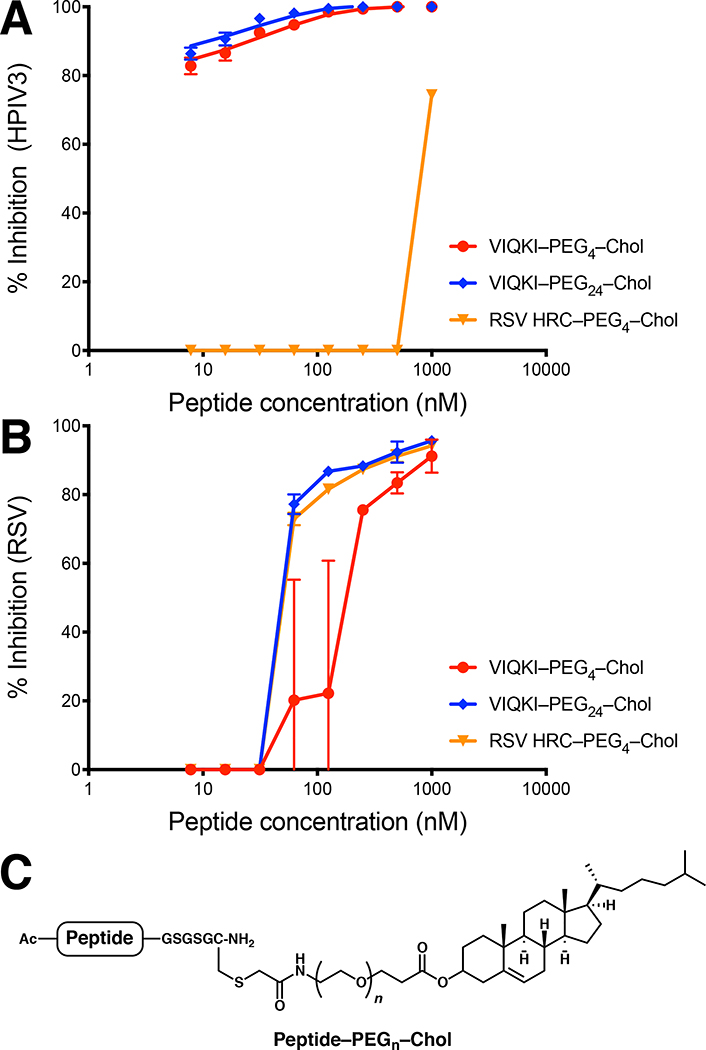
We prepared cholesterol-conjugated derivatives of HRC peptides (Fig. 3C) in order to explore the potential for antiviral activity in cell-based assays. Lipid conjugation has been previously shown to provide substantial increases in fusion-inhibitory potency for peptides based on viral fusion proteins, a trend that is attributed to lipid-promoted localization in membranes.20 A cell-cell fusion assay based on beta-galactosidase complementation was used to evaluate the inhibition of fusion mediated either by HPIV3 F co-expressed with HPIV3 HN or by RSV F (Table 2). Two VIQKI conjugates with different linker lengths (PEG4 and PEG24) were evaluated. As a control, we prepared an RSV HRC derivative with a PEG4 linker to a cholesterol moiety. Fusion mediated by HPIV3 F and HN was inhibited by the VIQKI conjugates but not the RSV HRC conjugate, even at the highest concentrations. VIQKI–PEG4–Chol and VIQKI–PEG24–Chol exhibited similar activities (IC50 ~ 20–100 nM). In contrast, fusion mediated by RSV F was inhibited by all three cholesterol-conjugated peptides with similar activities (IC50 ~ 1–3 μM). These results highlight the unique ability of VIQKI-derived peptides to inhibit fusion by both pediatric respiratory viruses.
Figure 3.
Inhibition of viral entry by VIQKI–cholesterol conjugates. Peptide activity against HPIV3 (A) or RSV (B) was determined by plaque reduction assay in infected cell monolayers. Cell monolayers were inoculated with 100 PFU of GFP-expressing HPIV3 or RFP-expressing RSV in the presence of various concentrations of peptides. After 90 min, overlay was added, and the plates were incubated at 37 °C for 24 h. Data were expressed as mean ± standard deviation (n = 3 separate experiments). (C) General structure of cysteine-linked peptide-cholesterol conjugates.
Table 2.
Inhibition of fusion mediated by HPIV3 or RSV F
| Peptide | HPIV3 IC50 (μM) | RSV IC50 (μM) |
|---|---|---|
| RSV HRC–PEG4–Chol | >10 | 3.05±1.27 |
| VIQKI–PEG4–Chol | 0.02±0.04 | 1.44±0.64 |
| VIQKI–PEG24–Chol | 0.10±0.08 | 3.31±1.65 |
Inhibition of HPIV3 and RSV entry
We determined the ability of cholesterol-conjugated VIQKI to inhibit HPIV3 or RSV infection using plaque reduction assays. We used recombinant HPIV3 that expresses enhanced green fluorescent protein (rHPIV3 CI-1-EGFP) and recombinant RSV that expresses red fluorescent protein (rRSV-RFP) so that the progression of infection could be quantified by fluorescence. As shown in Fig. 3A, the two VIQKI-derived conjugates blocked HPIV3 entry, but RSV HRC–PEG4–Chol was ineffective. For both VIQKI–PEG4–Chol and VIQKI–PEG24–Chol, >80% inhibition of plaque formation was observed at the lowest tested concentration (7.8 nM). In contrast, all three peptide conjugates were effective in the RSV plaque reduction assay (Fig. 3B), with VIQKI–PEG24–Chol (IC50 ~ 60 nM) somewhat more effective than VIQKI–PEG4–Chol (IC50 ~ 200 nM). This linker length trend is consistent with previous observations for HPIV3 and HIV.21,22 VIQKI–PEG24–Chol was as effective as RSV HRC–PEG4–Chol in this assay. It is possible that an RSV HRC peptide with a longer linker (e.g., RSV HRC–PEG24–Chol) would be more potent against RSV than either RSV HRC–PEG4–Chol or VIQKI–PEG24–Chol. However, given the very weak activity of RSV HRC-PEG4-Chol against HPIV3, it is very unlikely that simply lengthening the PEG linker would generate a potent inhibitor of HPIV3 entry. The key conclusion from the data in Fig. 3 is that an RSV HRC-derived agent is effective at inhibiting entry by only RSV, while VIQKI-derived agents are effective at inhibiting entry by both HPIV3 and RSV.
Inhibition of RSV infection in human airway epithelium
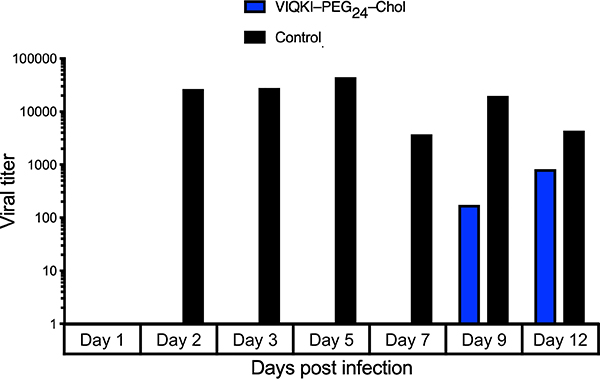
We and others have shown the value of assessing antiviral compounds in models that reflect the natural host. Such models are more likely to provide insights relevant to human medicine than other in vitro and in vivo animal models. We use human airway epithelium (HAE) cultures that form a multi-layered, polarized, differentiated tissue culture that closely resembles the airway epithelium in vivo. This model has been used to study RSV infection.23–28 Since VIQKI–PEG24–Chol demonstrated the best dual inhibitory activity in both fusion and plaque reduction assays, we tested the efficacy of this agent against RSV infection in HAE cultures (Fig. 4). HAE cultures were infected at the apical surface with rRSV-RFP in the presence of 10 μM of VIQKI–PEG24–Chol (blue) or untreated (black). After 90 min, the liquid from the apical surface, containing residual virus and peptide, was aspirated. The titer of virus emerging from the apical surface was measured at 1, 2, 3, 5, 7, 9, and 12 days post-infection. High viral titers were detected for the untreated cultures beginning at day 2 and were sustained until the study concluded at day 12. However, a single dose of VIQKI– PEG24–Chol led to no detectable virus until day 9, and the viral titers remained significantly lower than the untreated cultures at day 12. These observations strengthen the conclusion that use of the novel VIQKI sequence, which is based on the HPIV3 HRC sequence, can generate a potent inhibitor of RSV infection.
Figure 4.
Efficacy of VIQKI-PEG24-chol against RSV infection in human airway epithelial (HAE) cultures. HAE cultures were infected by applying 100 μl of EpiAirway medium containing 4,000 PFU of RSV to the apical surface for 90 min at 37 °C. The cultures were inoculated with a single dose of VIQKI–PEG24–Chol (10 μM, blue) or untreated control (black). The medium containing the inoculum was removed, cultures were placed at 37 °C and fed each day with 0.9 ml medium via the basolateral surface. Viruses were harvested by adding 200 μl medium per well to the HAE cultures’ apical surface and allowed to equilibrate for 30 min at 37 °C. The suspension was then collected, and viral titers were determined. Viral collection was performed sequentially with the same wells of cells on each day post infection.
Structural characterization of VIQKI bound to HRN domains of HPIV3 and RSV F
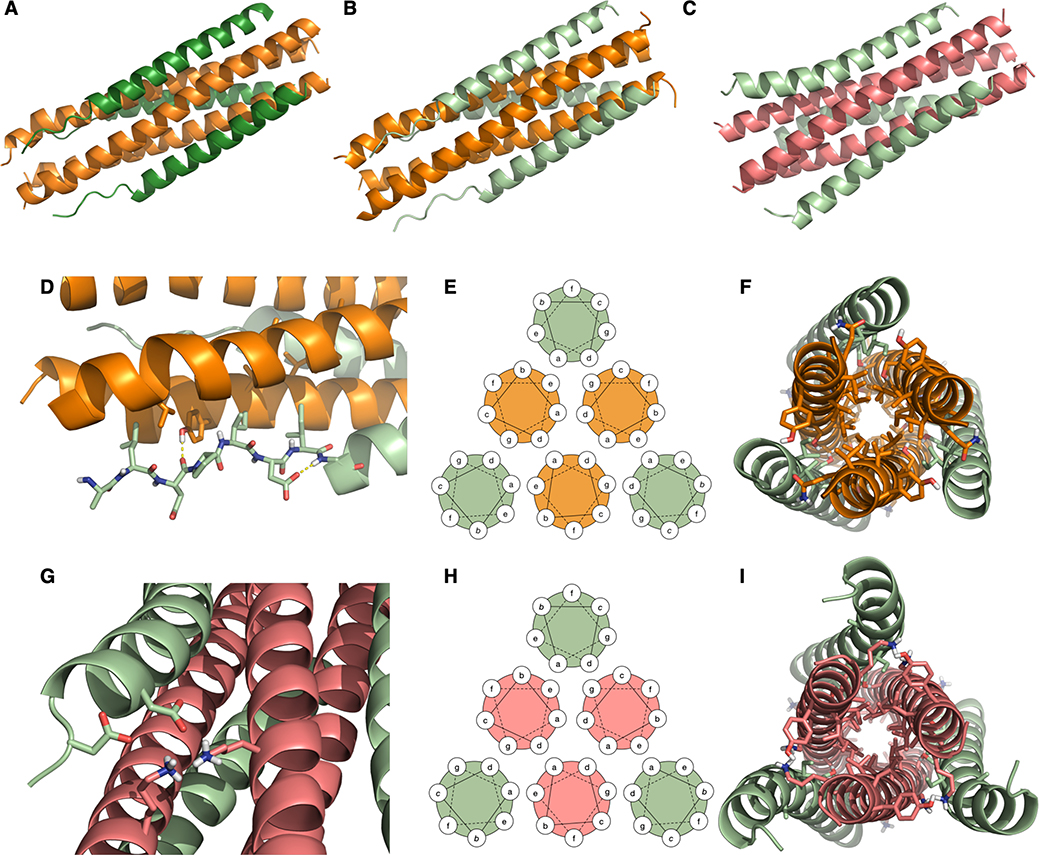
We determined co-crystal structures for VIQKI paired with either HPIV3 HRN (PDB 6NRO) or RSV HRN (PDB 6NTX). In addition, we determined the VI+HPIV3 HRN (PDB 6NYX) co-crystal structure. All three structures revealed six-helix bundle assemblies with helical HRN trimers at the core and three copies of VIQKI or VI on the periphery. The structures of VI or VIQKI co-assembled with the HRN domain of HPIV3 F are shown in Fig. 5A (1.85 Å resolution) and Fig. 5B (1.75 Å resolution), respectively. The structure of VIQKI co-assembled with the HRN domain of RSV is shown in Fig. 5C (2.20 Å resolution).
Figure 5.
Crystal structures of inhibitors bound to the HRN domains of HPIV3 and RSV F. (A) VI (dark green) + HPIV3 HRN (orange). (B) VIQKI (light green) + HPIV3 HRN (orange). (C) VIQKI (light green) + RSV HRN (salmon). (D) In the HPIV3 HRN+VIQKI co-assembly, the N-terminus of VIQKI adopts extended conformation with Leu451, Ile454, and Ile456 at the hydrophobic interface and Asp455 making an i,i+2 sidechain-to-backbone helix-capping interaction. (E) 6HB helical wheel diagram model. (F) Residues 457–484 follow the 6HB helical wheel model with a and d residues of the inhibitor interacting with a, d, e, and g residues of HPIV3 HRN. (G) In the RSV HRN+VIQKI co-assembly, the N-terminus of VIQKI adopts α-helical secondary structure with D452 and D455 engaged in salt bridge interactions with Lys191 and Lys196 of RSV HRN. (H) 6HB helical wheel diagram model. (I) Residues 463–484 follow 6HB helical wheel model with a and d residues of the inhibitor interacting with a, d, e, and g residues of RSV HRN.
The two structures containing HPIV3 HRN resemble the six-helix bundle portion within the structure of the postfusion form of the HPIV3 F ectodomain (PDB 1ZTM29). Each of the inhibitory peptides, VIQKI and VI, displays two regions with distinct secondary structure: the first 8 residues (corresponding to 449–456 of HPIV3 F, Fig. 5D), which display an extended conformation, and the remaining 28 residues (corresponding to 457–484 of HPIV3 F), which form an α-helix. The structure of VIQKI co-assembled with RSV HRN bears a general resemblance to the six-helix bundle assembly formed by RSV HRN and RSV HRC (PDB 1G2C30). The RSV HRC segment is largely α-helical in this assembly, with the first five residues (corresponding to 480– 484 of RSV F) adopting an extended conformation. Similarly, VIQKI is largely α-helical when co-assembled with RSV HRN (Fig. 5G); however, in this structure the three most N-terminal residues could not be resolved.
Polypeptide segments that engage in helix-bundle assemblies typically display a “heptad repeat” (HR) sequence pattern, with heptad positions designated by the letters abcdefg. The seven residues of a heptad can form approximately two α-helical turns. Sequences prone to helix-bundle assembly present aliphatic side chains (Leu, Ile or Val) at most a and d positions, which align to form a hydrophobic “stripe” in the helical conformation. In the six-helix bundle assemblies comprised of HRN and inhibitor peptides (Fig. 5E–F, 5H–I), the mostly hydrophobic residues at a and d positions of the inhibitors pack against mostly hydrophobic residues at a and d positions of either HPIV3 or RSV HRN. For the HRN segments, residues at e and g positions, which flank the stripe formed by the a and d residues, also contribute to the core of the six-helix bundle assembly. Some of the buried residues bear polar but uncharged side chains that form specific H-bonds within the assembly core. For example, in the VIQKI+HPIV3 HRN assembly, Ser470 (d) forms a H-bond with Gln162 (e), while Ser477 (d) forms H-bonds with Thr154 (d) from one molecule of HPIV3 HRN and with Asn155 (e) from a second molecule. In the VIQKI+RSV HRN structure, Ser477 forms a H-bond with Thr174 (d) from one molecule of RSV HRN and with Asn175 (e) from another molecule within the HRN trimer. Thr174 and Asn175 of RSV HRN align with Thr154 and Asn155 of HPIV3 HRN in the primary sequences. The Ser470 side chain of VIQKI does not appear to form any H-bond with RSV HRN.
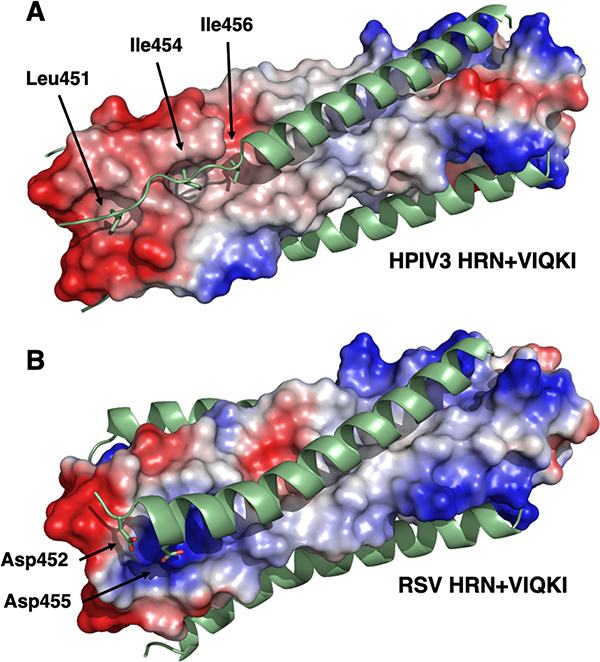
While residues 459–484 of VIQKI form an α-helix that engages the HRN trimers of both HPIV3 and RSV, as expected based on the heptad repeat motif, the co-crystal structures revealed that the N-terminus of VIQKI (residues 449–458) adopts markedly different conformations when engaged by the different HRN domains. In the assembly with HPIV3 HRN, Asp455 of VIQKI forms an intramolecular H-bond with the backbone amide N-H of Ser457. This i,i+2 interaction, characteristic of a helix-capping motif, is associated with the transition from the α-helical conformation displayed by most of the inhibitor to an extended conformation of residues near the N-terminus.31 The change in conformation positions hydrophobic residues Leu451, Ile454 and Ile456 of VIQKI towards hydrophobic cavities on the surface of the HPIV3 HRN trimer (Fig. 6A). In contrast, when co-assembled with RSV HRN, VIQKI displays a longer α-helix that starts at the fifth residue from the N-terminus. In this case, the backbone of the N-terminal segment of VIQKI curves away from the RSV HRN trimer. Ile454 and Ile456 of VIQKI project away from the RSV HRN trimer surface, in contrast to the intimate association of these Ile side chains with the HPIV3 HRN trimer surface. In place of these hydrophobic interactions, the N-terminal segment of VIQKI appears to engage in favorable Coulombic interactions with the RSV HRN trimer: the carboxylate side chains of Asp452 and Asp455 from one molecule of VIQKI form salt bridges with Lys191 from one molecule of RSV HRN and Lys196 of another molecule of RSV HRN (Fig. 6B). The “curving away” of the VIQKI backbone evident in Fig. 5C and 5I is necessary to allow these Coulombic contacts with the Lys residues of the RSV HRN trimer.
Figure 6.
Electrostatic potential maps of six-helix bundles assemblies for VIQKI with HPIV3 HRN (A) or RSV HRN (B). Locations of anionic (red), neutral (gray), or positive (blue) charge density are depicted.
Contributions of inhibitor N-terminal residue side chains to assembly stability
The crystal structures discussed above suggest that VIQKI adapts itself to two distinct viral targets, the HPIV3 HRN trimer or the RSV HRN trimer, by a profound conformational adjustment in the N-terminal segment of the inhibitor. In order to test this structure-based hypothesis, we used variable-temperature CD measurements to compare the stabilities of six-helix bundle assemblies formed by either HPIV3 HRN or RSV HRN with derivatives of VIQKI.
Initial studies involved truncated derivatives of VIQKI that lacked either the first three N-terminal residues (corresponding to residues 449–451 of the HPIV3 HRC domain) or the first eight N-terminal residues (corresponding to residues 449–456 of the HPIV3 HRC domain). Both of these truncated VIQKI peptides formed highly helical assemblies upon mixing with either HRN peptide, and each of the assemblies displayed a cooperative thermal transition, as monitored at 222 nm, that enabled us to determine a Tm,app value based on the transition midpoint (Table 3). In this case we calculated ΔTm,app values relative to VIQKI since we are comparing VIQKI analogues to VIQKI itself. Therefore, ΔTm,app‡ is the difference between Tm,app for the VIQKI derivative in question co-assembled with the HPIV3 HRN peptide and the Tm,app for the original VIQKI peptide co-assembled with the HPIV3 HRN peptide. ΔTm,app§ is the difference between Tm,app for the VIQKI derivative in question co-assembled with the RSV HRN peptide and the Tm,app for the original VIQKI peptide co-assembled with the RSV HRN peptide. Positive values indicate that the co-assembly of a particular VIQKI derivative with either HPIV3 or RSV HRN is more stable than that of VIQKI itself. Negative values indicate the co-assembly of a particular VIQKI derivative is less stable than that of VIQKI itself. Removal of the first three N-terminal residues, to generate VIQKI 452–484, caused very little change in the stability of the helical bundle assembly with either of the HRN peptides. In contrast, removal of the first eight N-terminal residues, to generate VIQKI 457–484, led to a profound decrease in the stabilities of both helix bundle assemblies. These latter observations are consistent with our hypothesis that contacts between the N-terminal segment of VIQKI and each HRN trimer are critical to the affinity with which the inhibitor peptide engages its viral target.
Table 3.
Apparent melting temperatures (Tm,app) of coassemblies formed between VIQKI variants and either HPIV3 HRN or RSV HRN
| HPIV3 HRN | RSV HRN | |||
|---|---|---|---|---|
| Inhibitor | Tm,app (°C) | ΔTm,appǂ | Tm,app (°C) | ΔTm,app§ |
| VIQKI | 88.6 | - | 64.3 | - |
| VIQKI 452–484 | 87.2 | −1.4 | 62.7 | −1.6 |
| VIQKI 457–484 | 63.8 | −24.8 | 36.8 | −27.5 |
| VIQKI V449A | >90.0 | >+1.4 | 66.5 | +2.2 |
| VIQKI L451A | 85.4 | −3.2 | 66.0 | +1.7 |
| VIQKI D452A | >90.0 | >+1.4 | 61.3 | −3.0 |
| VIQKI P453A | 89.9 | +1.3 | 67.6 | +3.3 |
| VIQKI I454A | 86.9 | −1.7 | 66.4 | +2.1 |
| VIQKI D455A | 82.1 | −6.5 | 60.2 | −4.1 |
| VIQKI I456A | 80.7 | −7.9 | 62.3 | −2.0 |
| VIQKI S457A | 87.4 | −1.2 | 67.9 | +3.6 |
ΔTm,appǂ = Tm,app (HPIV3 HRN+Inhibitor) − Tm,app (HPIV3 HRN+VIQKI)
ΔTm,app§ = Tm,app (RSV HRN+Inhibitor) − Tm,app (RSV HRN+VIQKI)
A second study involved an “alanine scan” of the N-terminal segment of VIQKI. Thus, a set of eight derivatives was prepared in which each of the N-terminal residues, in turn, was replaced by alanine. This experimental design allows one to assess the contribution of individual side chains to helix bundle stability (Table 3). Mutations D455A and I456A significantly destabilized the helix-bundle assembly with HPIV3 HRN. These observations are consistent with the structure of VIQKI assembled with HPIV3 HRN. The side chain carboxylate of Asp455 forms an H-bond with the backbone amide of Ser457, a helix-capping interaction that prevents extension of the helix toward the N-terminus of the inhibitor. This structure shows the side chain of Ile456 extended into a hydrophobic pocket on the surface of the HPIV3 HRN trimer. Trimming the native isobutyl side chain to a methyl group likely results in the loss of favorable hydrophobic interactions. The crystal structure suggests that the side chains of Leu451 and Ile454 might contribute similarly to helix-bundle stability, and the L451A and I454A mutation both caused decreases in Tm,app.
Among the helix-bundles formed with RSV HRN, the greatest destabilizations were observed for the mutations D452A and D455A. These observations are consistent with the structure of VIQKI assembled with RSV HRN, which indicates that the side chains of Asp452 and Asp455 form salt bridges with the side chains of Lys191 and Lys196 of RSV HRN. Changing either Asp to Ala would be expected to disrupt these favorable ionic interactions, which would account for the destabilization that results from either Ala mutation. Thus, even though the image in Fig. 5I suggests that the backbone of the N-terminal segment of VIQKI curves away from the binding groove on the RSV HRN trimer, this N-terminal segment retains thermodynamically favorable contacts with the RSV HRN trimer through the Asp-Lys contacts.
Discussion
Inhibition of the membrane fusion process that is required for viral entry is a promising strategy for combating infection.14, 32 We have previously described peptides derived from the HRC domain of HPIV3 F that inhibit fusion mediated by several paramyxoviral F proteins, including those of HPIV3, NiV and MeV.17–19 In those systems, we showed that conjugating a hydrophobic unit to the peptide significantly enhanced antiviral activity, presumably by promoting partitioning of the peptide inhibitor into the target membrane. The resulting lipopeptides were effective in vivo, blocking fusion by NiV in hamster and non-human primate models and by HPIV3 in a cotton rat model.18,19,33 Other lipopeptides have been safely and effectively used in humans for a variety of purposes, including as antibiotics, such as daptomycin, and as antivirals, such as a recent anti-HIV agent.34,35
The present study describes a substantial advance in the development of antiviral peptides with therapeutic potential: we have identified a new sequence based on the HRC domain of HPIV3 F, designated VIQKI, that displays potent inhibitory effects toward HPIV3 and the quite different pneumovirus RSV. The cholesterol-conjugated derivative VIQKI–PEG24–Chol potently blocked HPIV3 and RSV infection in plaque reduction assays and effectively reduced viral titers in mucociliary-differentiated HAE cultures that closely simulate in vivo human respiratory tissue. Because HPIV3 and RSV are major sources of pediatric morbidity and mortality, this new dual inhibitor represents an important step toward clinical application.
Crystal structures of six-helix bundle assemblies formed by VIQKI with the HRN domain of HPIV3 or RSV F show that the inhibitor binds along a groove formed by adjacent helices within an HRN domain trimer. These structures suggest that inhibitory activity results from VIQKI binding to a comparable surface within the extended prehairpin intermediate formed by the F trimer during the proposed membrane fusion mechanism. The prehairpin intermediate is a transient, high-energy state that is thought to precede the conformational transition to a more stable postfusion form that features a six-helix bundle assembly of HRN and HRC domains. The thermodynamic favorability of this structural transition is believed to drive the fusion of the viral envelope with the target cell membrane. Binding of VIQKI to the exposed HRN domain of the prehairpin intermediate would stabilize this otherwise transitory species, thereby blocking rearrangement to the postfusion form and preventing fusion of viral and cell membranes.
Our crystal structures provide a molecular explanation for the surprising ability of a peptide derived from the HRC domain of HPIV3 F to potently inhibit infection by RSV. VIQKI is largely helical in both structures, but the data show that VIQKI adapts itself to two different binding surfaces by accessing two distinct conformations in its N-terminal segment. The natural conformation, as seen in the postfusion form of the HPIV3 F ectodomain (PDB 1ZTM) and recapitulated in the assembly of VIQKI with HPIV3 HRN, features an extended backbone that allows HRC domain residues Ile454 and Ile456 to engage hydrophobic pockets on the surface of the HPIV3 HRN domain trimer. The thermodynamic importance of interactions involving Ile456 is indicated by our finding that the I456A modification of VIQKI significantly destabilizes the helical assembly formed with the HPIV3 HRN domain. The I454A modification, however, exerts only a small destabilizing effect.
The crystal structure of VIQKI co-assembled with the RSV HRN domain reveals that the helical portion of VIQKI extends further toward the N-terminus relative to VIQKI co-assembled with the HPIV3 HRN domain. This conformational change alters the set of residues that make contact with the RSV HRN trimer, relative to the HPIV3 HRN trimer. In the assembly with the RSV HRN domain, key N-terminal region contacts are made by Asp452 and Asp455 of VIQKI, which participate in salt bridges with Lys191 and Lys196 from the RSV HRN trimer. In the previously reported structure of RSV HRC co-assembled with RSV HRN (PDB 1G2C) Lys191 forms an H-bond with Ser491 of RSV HRC, while Lys196 interacts with Ser485 and Asp489 of RSV HRC.
Conclusions
We have designed a peptide, VIQKI, that binds to the HRN domains of both HPIV3 and RSV F. Modifying VIQKI by appending a cholesterol moiety, to generate VIQKI– PEG24–Chol, enabled potent inhibition of fusion mediated by both HPIV3 and RSV F, and the lipopeptide blocked infection by both viral pathogens. This dual-inhibitory activity is noteworthy given how different HPIV3 is from RSV. Our findings raise the prospect of a single therapeutic agent for treatment of acute respiratory tract infections in infants and children, without the need for identifying the pathogen. Structural insights on the mechanism of dual inhibition developed in this study should provide a basis for improvements in broad-spectrum antiviral activity.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH/NIAID R01AI114736 to AM and NIH/NIAID R01AI121349 to MP. VKO was funded in part by a Ruth L. Kirschstein National Research Service Award from the NIH (F32GM122263). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector was funded by an NIH–Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289). We thank Craig Bingman of the University of Wisconsin Crystallography Core facility for assistance and advice with x-ray data collection and processing and Shirley Chen for technical assistance.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: https://doi.org/10.1021/jacs.9b04615
Additional figures, synthetic procedures, and characterization data for all synthesized peptides, as well as protocols for fusion inhibition, plaque reduction, and thermal denaturation assays (PDF)
REFERENCES
- 1.Nair H; Nokes DJ; Gessner BD; Dherani M; Madhi SA; Singleton RJ; O’Brien KL; Roca A; Wright PF; Bruce N; Chandran A; Theodoratou E; Sutanto A; Sedyaningsih ER; Ngama M; Munywoki PK; Kartasasmita C; Simões EA; Rudan I; Weber MW; Campbell H Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. The Lancet 2010, 375, 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loughlin GM; Moscona A The cell biology of acute childhood respiratory disease: therapeutic implications. Pediatr. Clin. N. Am 2006, 53, 929–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Englund JA; Moscona A Paramyxoviruses: Parainfluenza viruses In Viral Infections of Humans Kaslow RA; Stanberry LR; Le Duc JW Eds., Springer, New York: 2014, 579–600. [Google Scholar]
- 4.Shi T; McAllister DA; O’Brien KL; Simoes EAF; Madhi SA; Gessner BD; Polack FP; Balsells E; Acacio S; Aguayo C; Alassani I; Ali A; Antonio M; Awasthi S; Awori JO; Azziz-Baumgartner E; Baggett HC; Baillie VL; Balmaseda A; Barahona A; Basnet S; Bassat Q; Basualdo W; Bigogo G; Bont L; Breiman RF; Brooks WA; Broor S; Bruce N; Bruden D; Buchy P; Campbell S; Carosone-Link P; Chadha M; Chipeta J; Chou M; Clara W; Cohen C; de Cuellar E; Dang D-A; Dash-Yandag B; Deloria-Knoll M; Dherani M; Eap T; Ebruke BE; Echavarria M; de Freitas Lázaro Emediato CC; Fasce RA; Feikin DR; Feng L; Gentile A; Gordon A; Goswami D; Goyet S; Groome M; Halasa N; Hirve S; Homaira N; Howie SRC; Jara J; Jroundi I; Kartasasmita CB; Khuri-Bulos N; Kotloff KL; Krishnan A; Libster R; Lopez O; Lucero MG; Lucion F; Lupisan SP; Marcone DN; McCracken JP; Mejia M; Moisi JC; Montgomery JM; Moore DP; Moraleda C; Moyes J; Munywoki P; Mutyara K; Nicol MP; Nokes DJ; Nymadawa P; da Costa Oliveira MT; Oshitani H; Pandey N; Paranhos-Baccalà G; Phillips LN; Picot VS; Rahman M; Rakoto-Andrianarivelo M; Rasmussen ZA; Rath BA; Robinson A; Romero C; Russomando G; Salimi V; Sawatwong P; Scheltema N; Schweiger B; Scott JAG; Seidenberg P; Shen K; Singleton R; Sotomayor V; Strand TA; Sutanto A; Sylla M; Tapia MD; Thamthitiwat S; Thomas ED; Tokarz R; Turner C; Venter M; Waicharoen S; Wang J; Watthanaworawit W; Yoshida L-M; Yu H; Zar HJ; Campbell H; Nair H; RSV Global Epidemiology Network. Global, Regional, and National Disease Burden Estimates of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children in 2015: a Systematic Review and Modelling Study. Lancet 2017, 390 (10098), 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Committee on Infectious Diseases and Bronchiolitis Guidelines Committee. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 2014, 134, e620–e638. [DOI] [PubMed] [Google Scholar]
- 6.Harrison SC Viral membrane fusion. Nat. Struct. Mol. Biol 2008, 15, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A; Dutch RE Paramyxovirus fusion and entry: Multiple paths to a common end. Viruses 2012, 4, 613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kielian M Mechanisms of virus membrane fusion proteins. Ann. Rev. Virol 2014, 1, 171–189. [DOI] [PubMed] [Google Scholar]
- 9.Lambert DM; Barney S; Lambert AL; Guthrie K; Medinas R; Davis DE; Bucy T; Erickson J; Merutka G; Petteway SR Peptides From Conserved Regions of Paramyxovirus Fusion (F) Proteins Are Potent Inhibitors of Viral Fusion. Proc. Natl. Acad. Sci USA 1996, 93 (5), 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wild C; Dubay JW; Greenwell T; Baird T; Oas TG; McDanal C; Hunter E; Matthews T Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp4l to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 1994, 91 (26), 12676–12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Q; Compans RW Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 1996, 223, 103–112. [DOI] [PubMed] [Google Scholar]
- 12.Rapaport D; Ovadia M; Shai Y A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995, 14, 5524–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolarinwa O; Zhang M; Mulry E; Lu M; Cai J Sulfono-ɣ-AA modified peptides that inhibit HIV-1 fusion. Org. Biomol. Chem 2018, 16 (42), 7878–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews T; Salgo M; Greenberg M; Chung J; DeMasi R; Bolognesi D Enfuvirtide: The first therapy to inhibit the entry of HIV-1 into host CD4 lymophocytes. Nat. Rev. Drug Discov 2004, 3 (3), 215–225. [DOI] [PubMed] [Google Scholar]
- 15.Porotto M; Carta P; Deng Y; Kellogg GE; Whitt M; Lu M; Mungall BA; Moscona A Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J. Virol 2007, 81 (19), 10567–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieu C; Huey D; Jurgens E; Welsch JC; DeVito I; Talekar A; Horvat B; Niewiesk S; Moscona A; Porotto M Prevention of measles virus infection by intranasal delivery of fusion inhibitor peptides. J. Virol 2015, 89 (2), 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueira TN; Palermo LM; Veiga AS; Huey D; Alabi CA; Santos NC; Welsch JC; Mathieu C; Horvat B; Niewiesk S; Moscona A; Castanho MARB; Porotto M In vivo efficacy of measles virus fusion protein-derived peptides is modulated by the properties of self-assembly and membrane residence. J. Virol 2017, 91 (1), e01554–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathieu C; Augusto MT; Niewiesk S; Horvat B; Palermo LM; Sanna G; Madeddu S; Huey D; Castanho MARB; Porotto M; Santos NC; Moscona A Broad spectrum antiviral activity for paramyxoviruses is modulated by biophysical properties of fusion inhibitory peptides. Sci. Rep 2017, 7, 43610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porotto M; Rockx B; Yokoyama CC; Talekar A; DeVito I; Palermo LM; Liu J; Cortese R; Lu M; Feldmann H; Pessi A; Moscona A Inhibition of Nipah virus infection in vivo: Targeting an early stage paramyxovirus fusion activation during viral entry. PLoS Pathog. 2010, 6 (10), e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porotto M; Yokoyama CC; Palermo LM; Mungall B; Aljofan M; Cortese R; Pessi A; Moscona A Viral entry inhibitors targeted to the membrane site of action. J. Virol 2010, 84 (13), 6760–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pessi A; Langella A; Capitò E; Ghezzi S; Vicenzi E; Poli G; Ketas T; Mathieu C; Cortese R; Horvat B; Moscona A; Porotto M A general strategy to endow natural fusion protein-derived peptides with potent antiviral activity. PLoS ONE 2012, 7 (5), e36833–e36839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augusto MT; Hollmann A; Troise F; Veiga AS; Pessi A; Santos NC Lipophilicity is a key factor to increase the antiviral activity of HIV neutralizing antibodies. Colloids Surf. B Biointerfaces 2017, 152, 311–316. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L; Bukreyev A; Thompson CI; Watson B; Peeples ME; Collins PL; Pickles RJ Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol 2005, 79 (2), 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellow TE; Murphy PC; Carson JL; Noah TL; Zhang L; Pickles RJ The effect of respiratory syncytial virus on chemokine release by differentiated airway epithelium. Exp. Lung Res 2004, 30, 43–57. [DOI] [PubMed] [Google Scholar]
- 25.Palermo LM; Porotto M; Yokoyama CC; Palmer SG; Mungall BA; Greengard O; Niewiesk S; Moscona A Human parainfluenza virus infection of the airway epithelium: Viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol 2009, 83 (13), 6900–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palermo LM; Uppal M; Skrabanek L; Zumbo P; Germer S; Toussaint NC; Rima BK; Huey D; Niewiesk S; Porotto M; Moscona A Features of circulating parainfluenza virus required for growth in human airway. mBio 2016, 7 (2), e00235–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer SG; Porotto M; Palermo LM; Cunha LF; Greengard O; Moscona A mBio 2012, 3 (3), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iketani S; Shean RC; Ferren M; Makhsous N; Aquino DB; Georges, des A; Rima B; Mathieu C; Porotto M; Moscona A; Greninger AL Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. mBio 2018, 9 (4), 1545–19. [Google Scholar]
- 29.Yin H-S; Paterson RG; Wen X; Lamb RA; Jardetzky TS Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 2005, 102 (26), 9288–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X; Singh M; Malashkevich VN; Kim PS Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 2000, 97 (26), 14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano L; Fersht AR Capping and α-helix stability. Nature 1989, 342 (6247), 296–299. [DOI] [PubMed] [Google Scholar]
- 32.Moore JP; Doms RW The entry of entry inhibitors: A fusion of science and medicine. Proc. Natl. Acad. Sci. USA 2003, 100 (19), 10598–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathieu C; Porotto M; Figueira TN; Horvat B; Moscona A Fusion inhibitory lipopeptides engineered for prophylaxis of Nipah virus in primates. J. Infect. Dis 2018, 218 (2), 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steenbergen JN; Alder J; Thorne GM; Tally FP Daptomycin: a Lipopeptide Antibiotic for the Treatment of Serious Gram-Positive Infections. Journal of Antimicrobial Chemotherapy 2005, 55 (3), 283–288. [DOI] [PubMed] [Google Scholar]
- 35.Quinn K; Traboni C; Penchala SD; Bouliotis G; Doyle N; Libri V; Khoo S; Ashby D; Weber J; Nicosia A; Cortese R; Pessi A; Winston A A First-in-Human Study of the Novel HIV-Fusion Inhibitor C34PEG4-Chol. Sci. Rep 2017, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.